Exploring the Molecular Regulatory Mechanisms of Planktonic Larvae Development in Mytilus coruscus Using Weighted Gene Co-expression Network Analysis and Time-series Differential Analysis
-
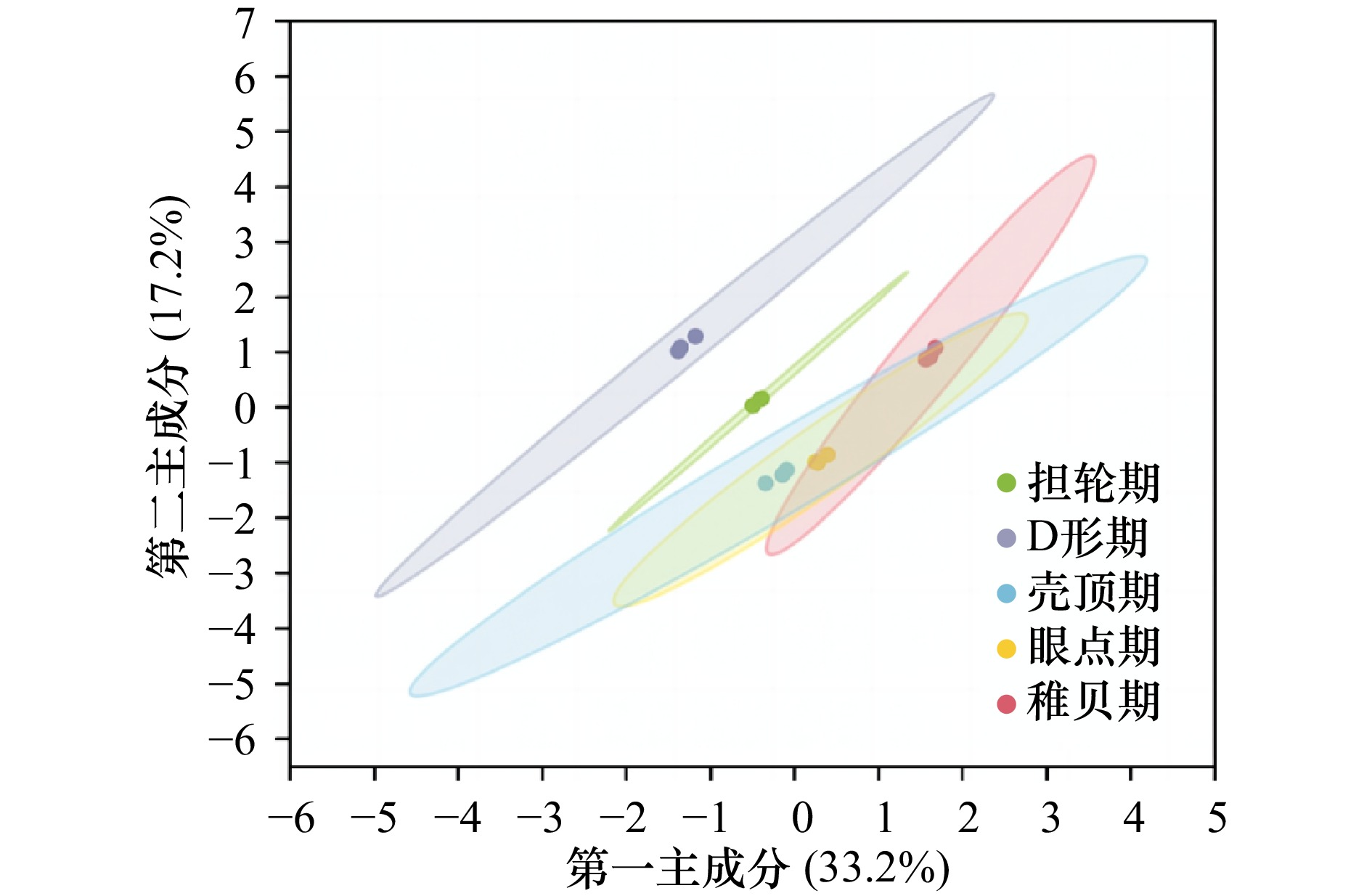
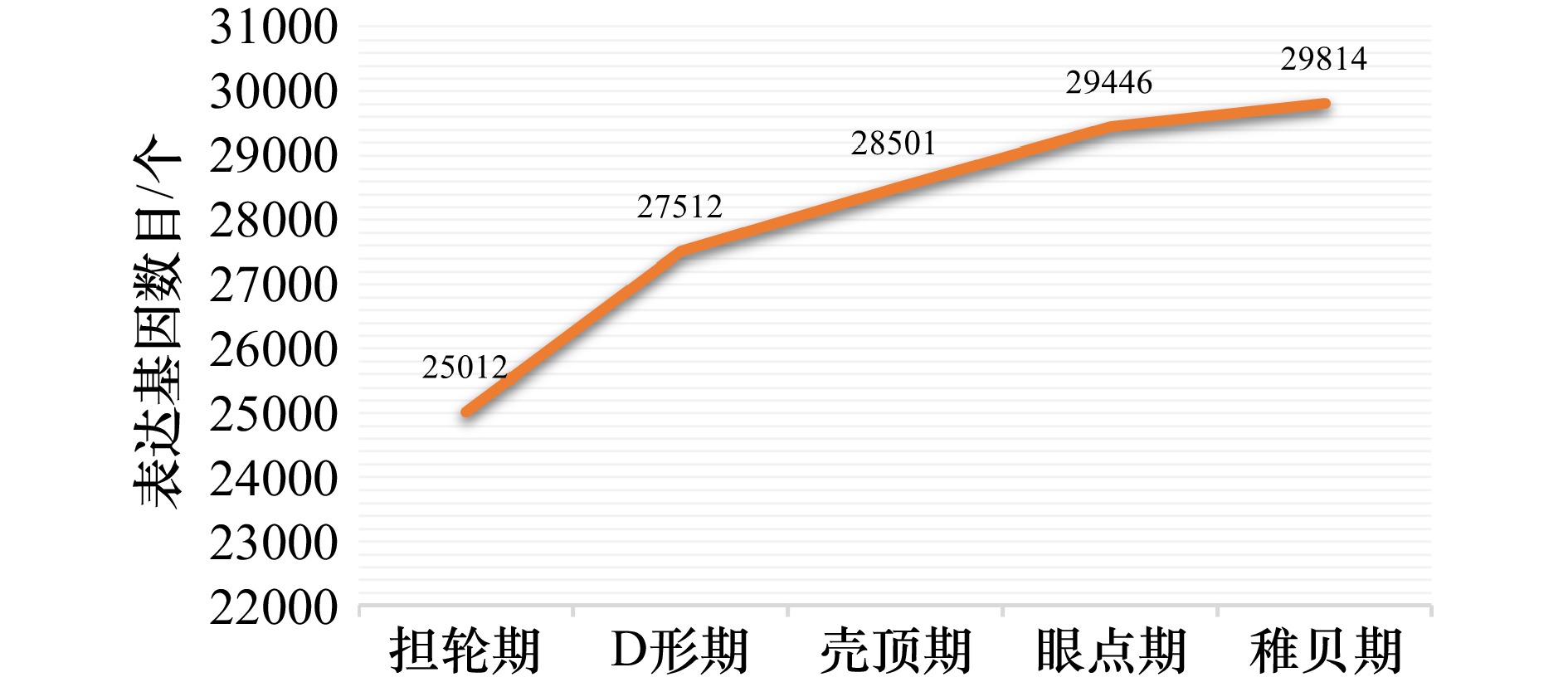
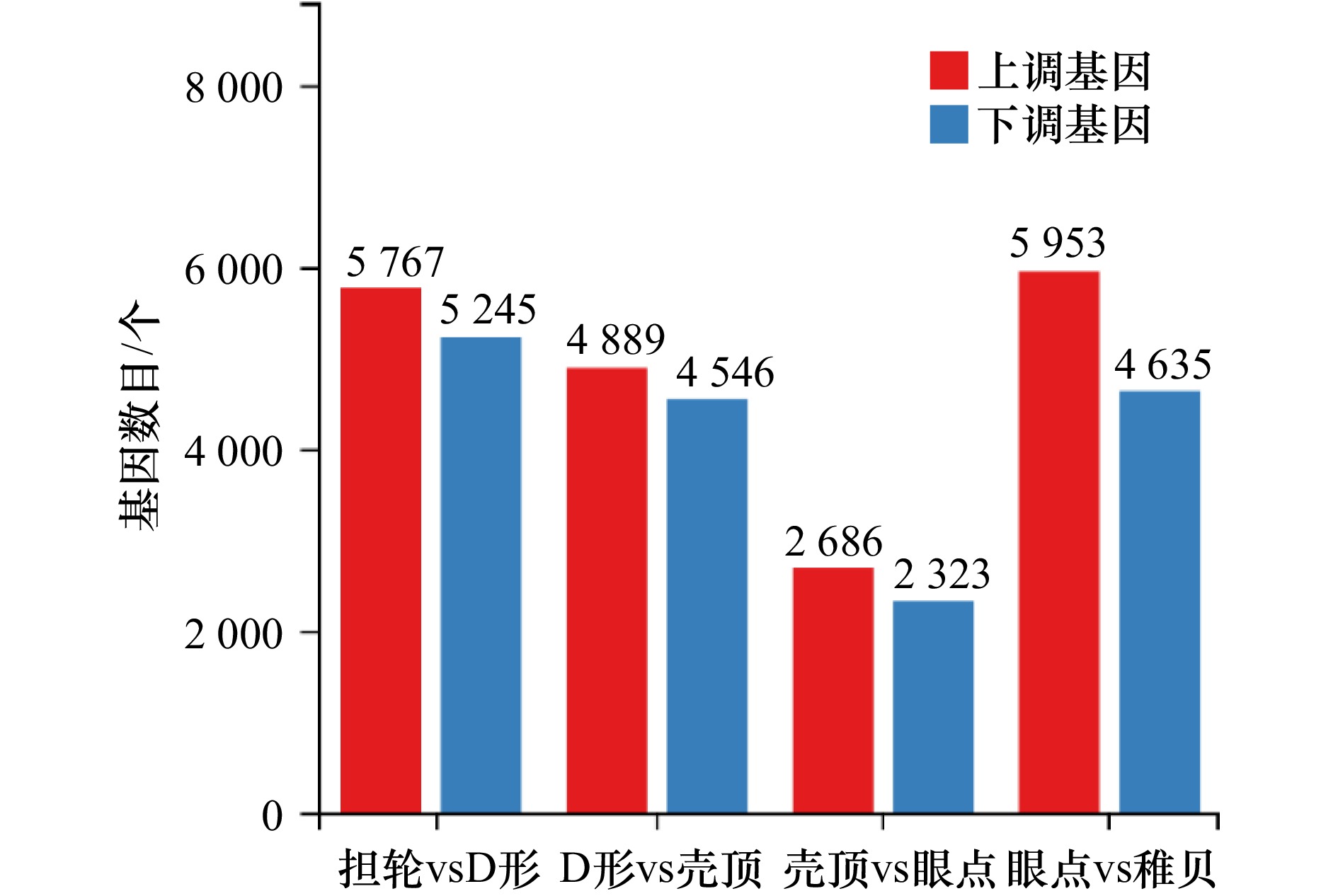
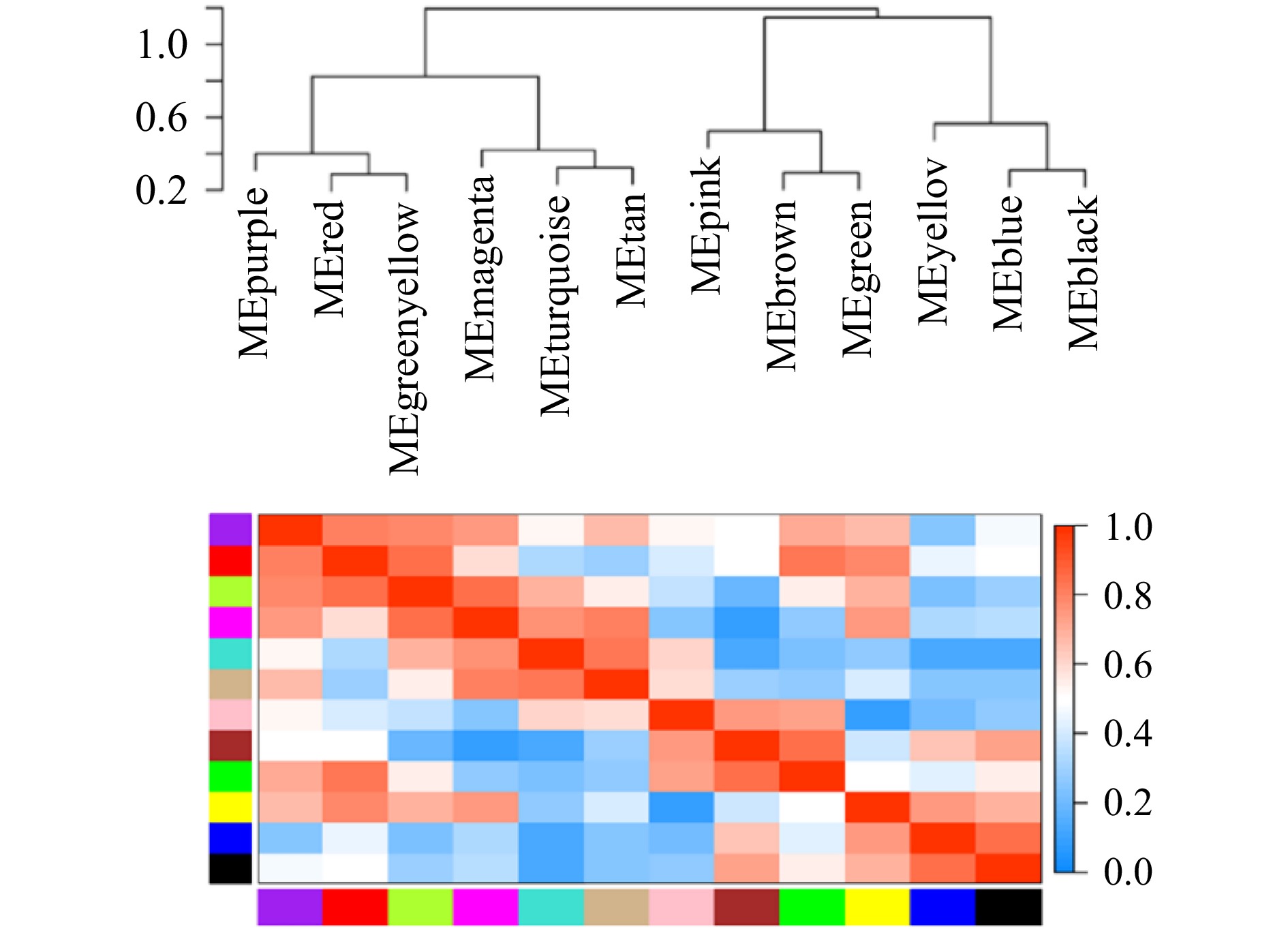
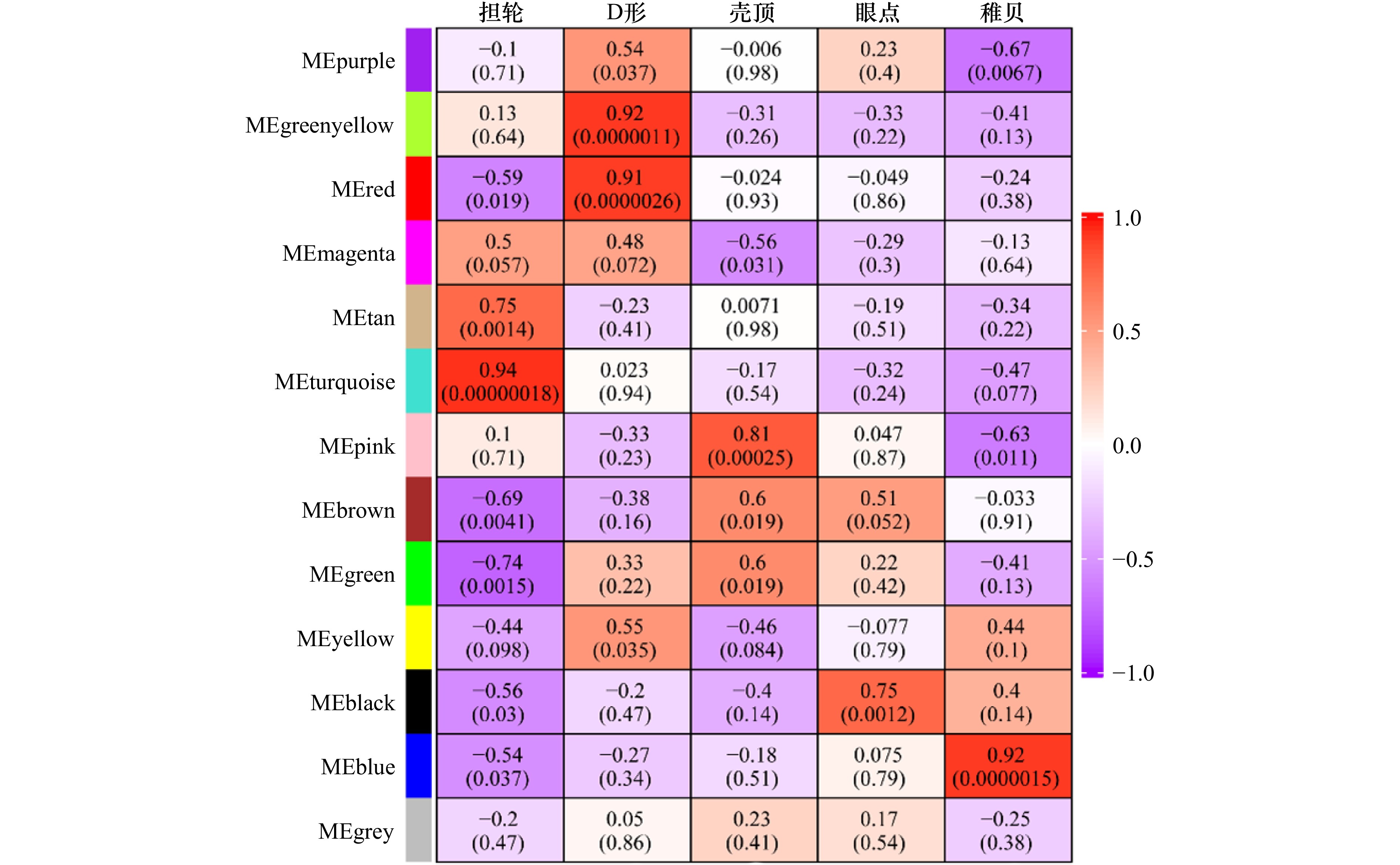
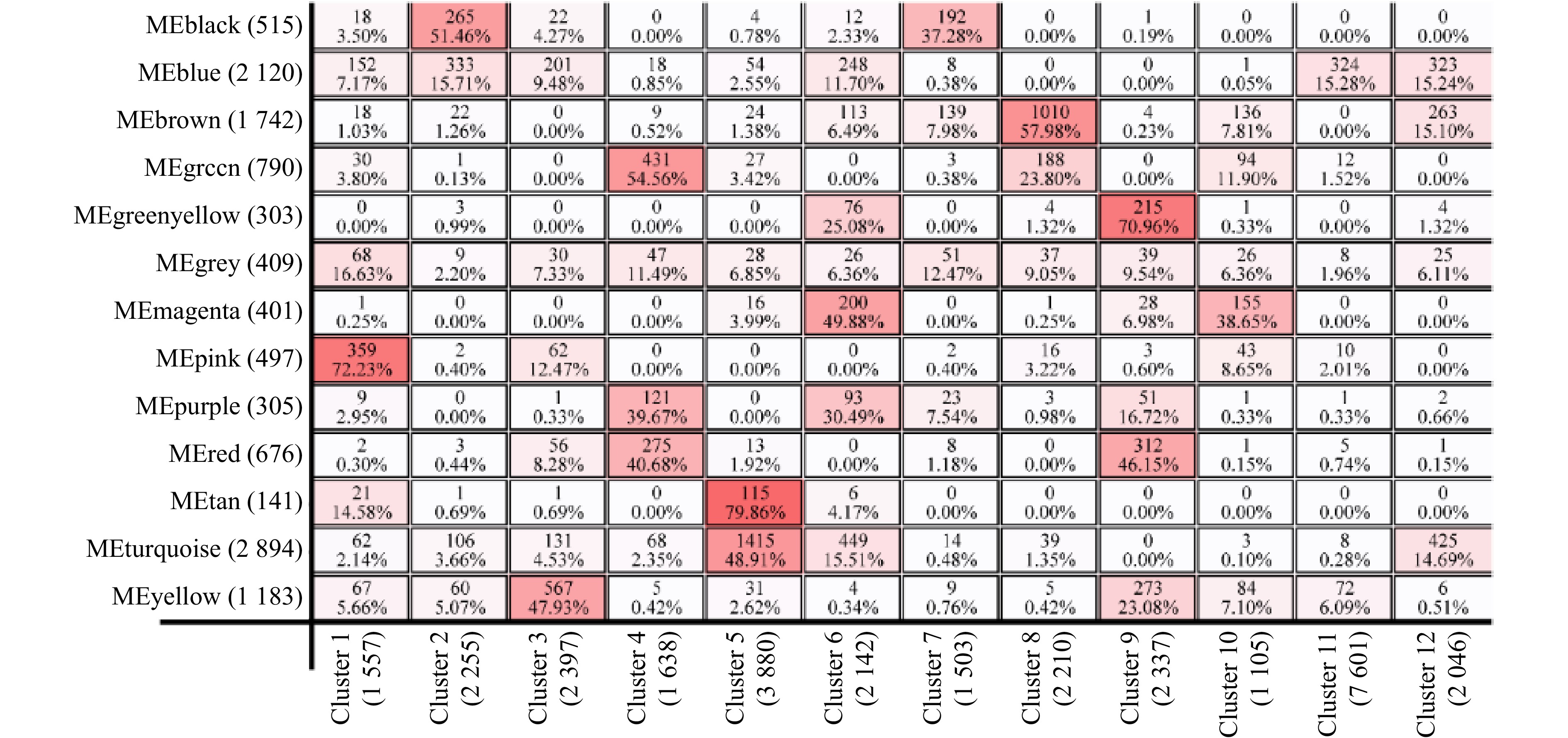
摘要: 厚壳贻贝(Mytilus coruscus)味道鲜美,富含多种营养物质,是我国沿海重要的经济贝类之一。厚壳贻贝育苗的关键时期在其幼虫变态发育阶段,而幼虫发育是一个动态且复杂的过程,有大量基因的参与以及各种复杂生物学过程共同作用。本研究基于高通量测序技术,对厚壳贻贝幼虫发育的5个关键时期(担轮期、D形期、壳顶期、眼点期、稚贝期)样本进行高通量转录组测序,共筛选出20 894个差异基因。对差异基因进行加权基因共表达网络以及时序差异分析,筛选出6个符合特定时序发育模式的关键子模块,共鉴定出2 395个基因。分别对各子模块内基因进行GO富集分析和蛋白网络互作分析,筛选出30个与厚壳贻贝生长发育过程相关的枢纽基因,包括Fen1、Ndufab1b、Ndufs8a、Pcan、Rnaseh2a;Cdh1、Cacng4b、Cav1、Blm、Ryr1a;Mars1、Cdc42、Aasdh、Apoba、Cav1;Kif11、Cdc20、Ubc、Kif23、Cdc6;Ubc、Rps16、Rpl23、Rpsa、Rps27a;Cdc20、Setd2、Ssrp1a、Cav1、Rab8a。这些基因在厚壳贻贝幼虫发育过程的主要参与调控DNA复制和细胞分裂、线粒体与核糖体功能、蛋白质合成等。本研究在转录组水平上探索了厚壳贻贝浮游幼虫发育过程调控的分子机理,对研究厚壳贻贝功能基因,以及后续培育更优表型性状厚壳贻贝新品种具有重要的理论指导意义。Abstract: Mytilus coruscus is renowned for its delicious taste and rich nutritional content, making it one of the significant economic shellfish along the coast of China. The critical period for M. coruscus seedling cultivation lies in its larval metamorphosis stage. However, the larval development of M. coruscus is a dynamic and complex process, involving the participation of numerous genes and various intricate biological processes. In this study, we based on high-throughput transcriptome sequencing technology, conducted transcriptome sequencing on larval samples of M. coruscus at five key developmental stages (Trocophore, D-veliger, Umbo, Pediveliger, and Post-larvae), identifying a total of 20 894 differentially expressed genes. Weighted gene co-expression network analysis and time-course difference analysis were performed on the differentially expressed genes, followed by joint analysis. Six key submodules conforming to specific temporal developmental patterns were selected, identifying a total of 2 395 genes. Gene Ontology enrichment analysis and protein network interaction analysis were conducted on genes within each submodule. Thirty hub genes related to the growth and development process of M. coruscus were identified, such as Fen1、Ndufab1b、Ndufs8a、Pcan、Rnaseh2a; Cdh1、Cacng4b、Cav1、Blm、Ryr1a; Mars1、Cdc42、Aasdh、Apoba、Cav1; Kif11、Cdc20、Ubc、Kif23、Cdc6; Ubc、Rps16、Rpl23、Rpsa、Rps27a; Cdc20、Setd2、Ssrp1a、Cav1、Rab8a. They playing major regulatory roles in larval development, including regulation of DNA replication and cell division processes, mitochondrial and ribosomal functions, and protein synthesis processes. This study explores the molecular mechanisms underlying the regulation of M. coruscus larval development at the transcriptome level, providing important theoretical guidance for studying functional genes in M. coruscus and subsequent molecular breeding practices aimed at cultivating new varieties with superior phenotypic traits.
-
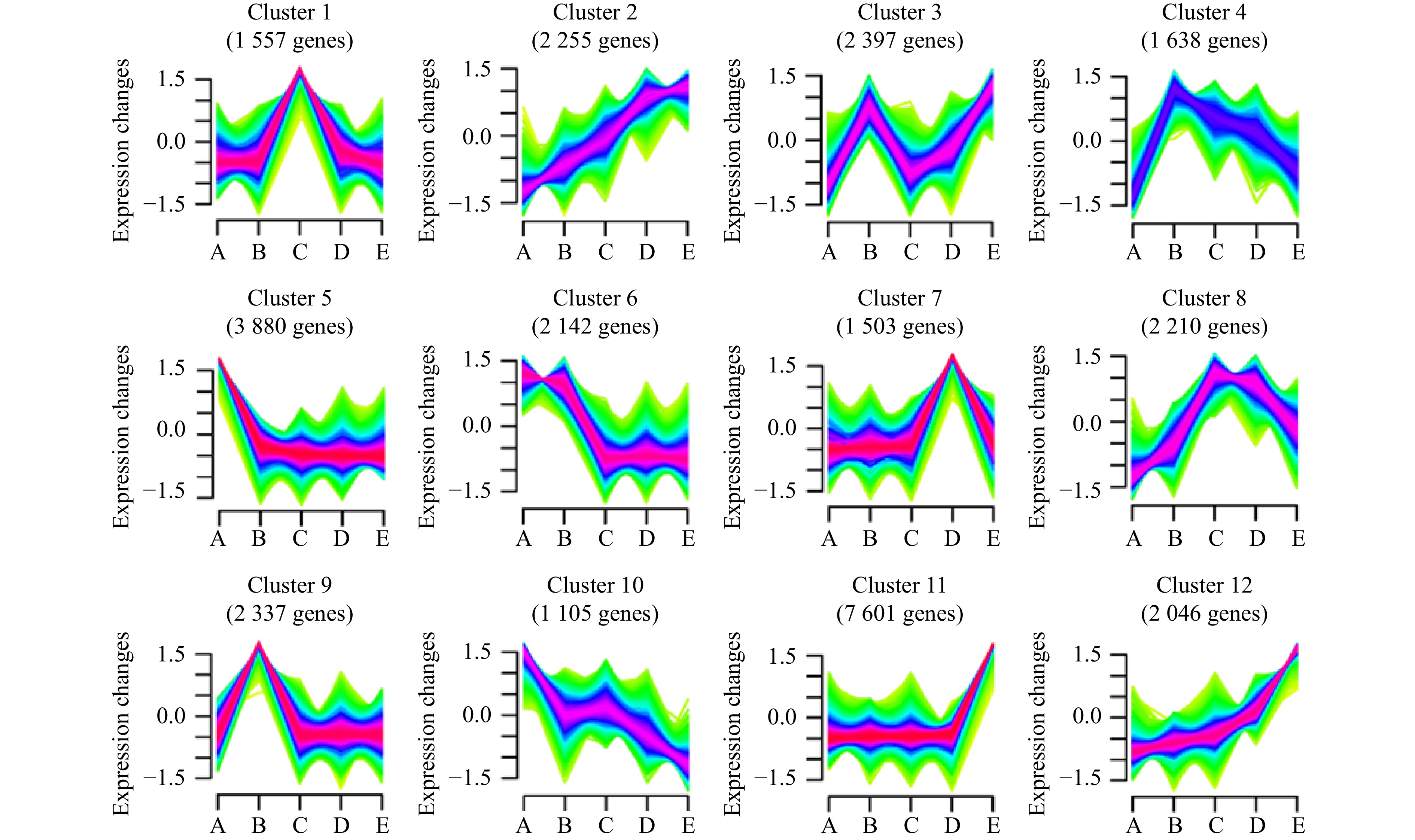
图 13 6个子模块中基因的GO富集分析
a为Cluster1-MEpink,b为Cluster2-MEblack,c为Cluster4-MEgreen,d为Cluster5-MEtan,e为Cluster8-MEbrown,f为Cluster9-MEgreenyellow.
Fig. 13 GO enrichment analysis of genes in six submodules
a is Cluster1-MEpink, b is Cluster2-MEblack, c is Cluster4-MEgreen, d is Cluster5-MEtan, e is Cluster8-MEbrown, f is Cluster9-MEgreenyellow.
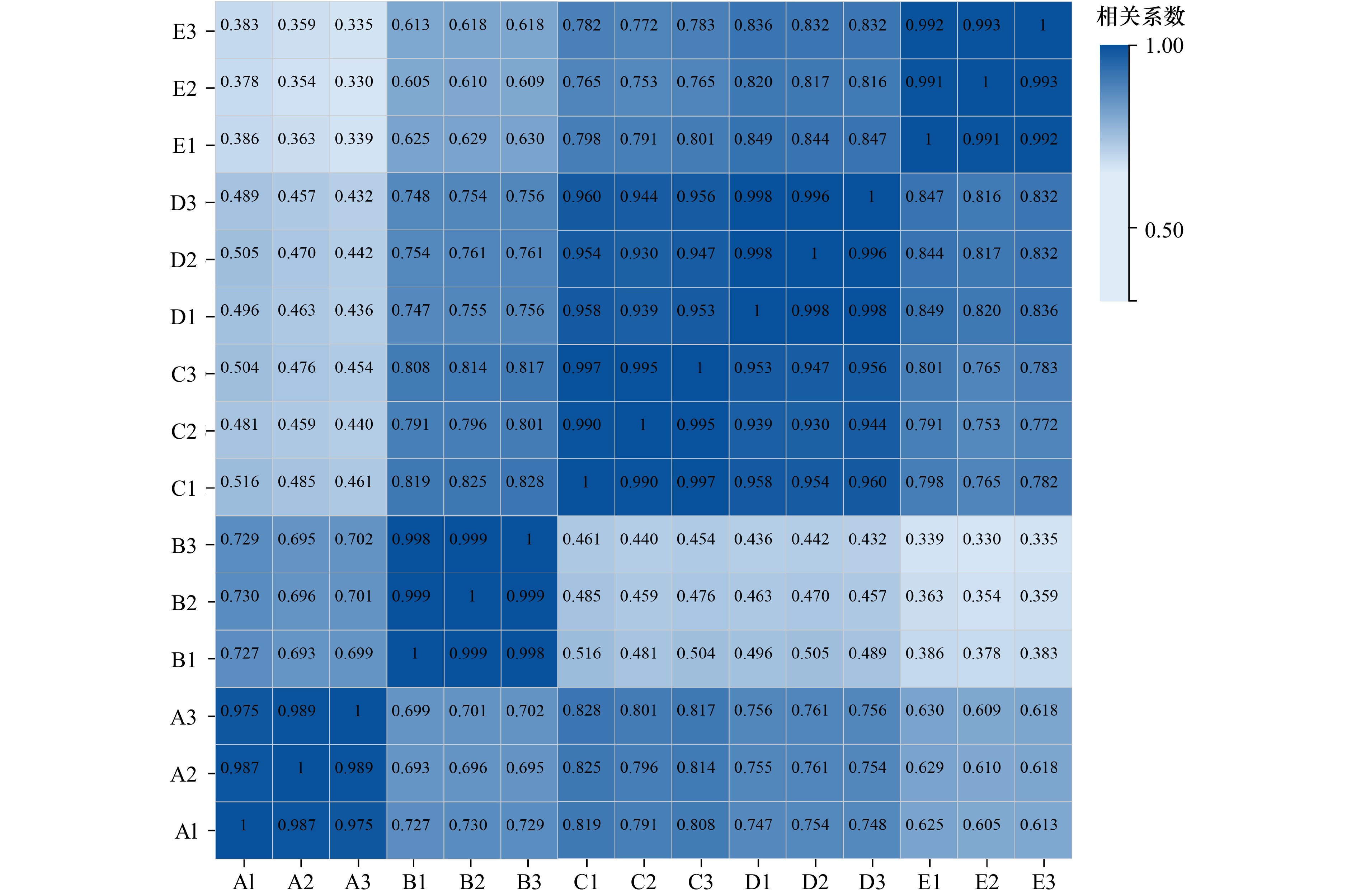
表 1 转录组数据质控结果统计
Tab. 1 Statistics of transcriptome data quality control results
样本
Sample原始读长度Raw Reads (M) 高质量读
长度Clean Reads (M)碱基正确率在99%的占比Q20 (%) 碱基正确率在99.9%的占比Q30 (%) 能定位到基因组上的
高质量读长
数目占比Mapped Ratio (%)A1 43.82 42.16 96.53 91.37 96.21 A2 43.82 42.28 96.73 91.85 96.49 A3 45.57 42.42 97.01 92.51 93.08 B1 45.57 43.37 96.74 91.90 95.16 B2 43.82 41.99 96.78 91.91 95.82 B3 43.82 42.16 96.84 92.09 96.22 C1 43.82 42.10 96.72 91.82 96.08 C2 43.82 42.02 96.83 92.09 95.88 C3 43.82 42.23 96.69 91.73 96.37 D1 43.82 42.29 96.54 91.35 96.50 D2 43.82 42.41 97.48 93.32 96.78 D3 43.82 42.30 97.54 93.44 96.53 E1 43.82 42.19 97.24 92.81 96.28 E2 43.82 42.10 97.21 92.71 96.07 E3 43.82 42.03 97.19 92.7 95.91 平均值 44.05 42.27 96.94 92.24 95.96 注:A1−A3为担轮期、B1−B3为D形期、C1−C3为壳顶期、D1−D3为眼点期、E1−E3为稚贝期;M即Million,代表每百万条。 Notes: A1-A3 is Trocophore, B1-B3 is D-veliger, C1-C3 is Umbo, D1-D3 is Pediveliger, E1-E3 is Post-larvae; M is Million. 表 2 GO条目富集统计表
Tab. 2 Statistics of enrichment GO terms
模块 Moudle 显著富集GO条目 分子功能(MF) 细胞组分(CC) 生物过程(BP) Cluster1-MEpink 214 51 54 109 Cluster2-MEblack 50 26 16 8 Cluster4-MEgreen 37 10 1 26 Cluster5-MEtan 47 11 10 26 Cluster8-MEbrown 201 46 41 114 Cluster9-MEgreenyellow 50 24 13 13 表 3 表达基因中的转录因子分布情况统计
Tab. 3 Statistics on the distribution of transcription factors among expressed genes
基因家族
TF familyCluster1-
MEpinkCluster2-
MEblackCluster4-
MEgreenCluster5-
MEtanCluster8-
MEbrownCluster9-
MEgreenyellowARID 0 2 1 1 4 0 ETS 0 0 0 1 0 1 Fork 0 0 0 1 1 0 HMG 1 0 2 1 1 0 Homeobox 1 8 7 1 7 4 MBD 1 0 0 0 0 1 PAX 1 0 0 0 0 0 T-box 0 2 1 1 1 1 THAP 0 0 1 0 1 0 ZBTB 0 1 0 4 0 0 注:第2−7列代表每个子模块中该转录因子表达基因的个数。 Notes: columns 2-7 in the table represent the number of genes expressed by this transcription factor in each sub-module. -
[1] 张玺, 齐钟彦. 贝类学纲要[M]. 北京: 科学出版社, 1961.(第27条、第29条文献在正文里未被引用,请确认)Zhang Xi, Qi Zhongyan. Outline of Conchology[M]. Beijing: Science Press, 1961. (查阅网上资料, 未找到本条文献英文信息, 请确认) [2] 张永普, 郑洁, 王一农. 浙南岛屿岩相潮间带贻贝类的生态特点[J]. 海洋湖沼通报, 2000(3): 24−28. doi: 10.3969/j.issn.1003-6482.2000.03.006Zhang Yongpu, Zheng Jie, Wang Yinong. Ecological characteristics of the intertidal mussels of the islands south of Zhejiang[J]. Transactions of Oceanology and Limnology, 2000(3): 24−28. doi: 10.3969/j.issn.1003-6482.2000.03.006 [3] 张义浩. 浙江沿海贻贝种类形态比较研究[J]. 渔业经济研究, 2009(2): 14−20.Zhang Yihao. Study on shape comparison of mussel species in Zhejiang coast[J]. Fisheries Economy Research, 2009(2): 14−20. [4] 叶莹莹, 徐梅英, 吴常文. 几种环境因子对厚壳贻贝浮游幼虫生长与存活的影响[J]. 浙江海洋学院学报(自然科学版), 2011, 30(4): 292−296.Ye Yingying, Xu Meiying, Wu Changwen. Influences of some environmental factors on growth and survival of Mytilus coruscus gould larvae[J]. Journal of Zhejiang Ocean University(Natural Science), 2011, 30(4): 292−296. [5] 王朝新. 厚壳贻贝苗种规模化繁育技术[J]. 现代农业科技, 2021(17): 208−210.Wang Chaoxin. Large-scale breeding technology of thick shell mussel[J]. Modern Agricultural Science and Technology, 2021(17): 208−210. (查阅网上资料, 未找到本条文献英文信息, 请确认) [6] 颜成瑞. 厚壳贻贝Hox基因家族结构与表达模式的初步探究[D]. 舟山: 浙江海洋大学, 2022.Yan Chengrui. Preliminary exploration of Hox Gene family structure and expression pattern in Mytilus coruscus[D]. Zhoushan: Zhejiang Ocean University, 2022. [7] 顾忠旗, 倪梦麟, 范卫明. 厚壳贻贝胚胎发育观察[J]. 安徽农业科学, 2010, 38(32): 18213−18215. doi: 10.3969/j.issn.0517-6611.2010.32.089Gu Zhongqi, Ni Menglin, Fan Weiming. Observation on embryonic development of Mytilus coruscus[J]. Journal of Anhui Agricultural Sciences, 2010, 38(32): 18213−18215. doi: 10.3969/j.issn.0517-6611.2010.32.089 [8] 徐嘉康, 王劲松, 方怡涵, 等. 厚壳贻贝肠道细菌的生物被膜对其幼虫和稚贝附着的影响[J]. 海洋学报, 2021, 43(9): 81−91.Xu Jiakang, Wang Jinsong, Fang Yihan, et al. Effects of intestinal bacterial biofilms on settlement process of larvae and plantigrades in Mytilus coruscus[J]. Haiyang Xuebao, 2021, 43(9): 81−91. [9] 林欣. 四指马鲅(Eleutheronema tetradactylum)应对低氧胁迫的组织影响以及鳃转录组学分析研究[D]. 大连: 大连海洋大学, 2023.Lin Xin. Tissue effects and gill transcriptomic analysis of Eleutheronema tetradactylum in response to hypoxia stress[D]. Dalian: Dalian Ocean University, 2023. (查阅网上资料, 未找到本条文献英文信息, 请确认) [10] Brown D D. Gene expression in eukaryotes[J]. Science, 1981, 211(4483): 667−674. doi: 10.1126/science.6256857 [11] Cramer P. Organization and regulation of gene transcription[J]. Nature, 2019, 573(7772): 45−54. doi: 10.1038/s41586-019-1517-4 [12] Chen Yuxin, Chen Yongsheng, Shi Chunmei, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data[J]. GigaScience, 2018, 7(1): gix120. [13] Kim D, Langmead B, Salzberg S L. HISAT: a fast spliced aligner with low memory requirements[J]. Nature Methods, 2015, 12(4): 357−360. doi: 10.1038/nmeth.3317 [14] Langmead B, Salzberg S L. Fast gapped-read alignment with Bowtie 2[J]. Nature Methods, 2012, 9(4): 357−359. doi: 10.1038/nmeth.1923 [15] Li Bo, Dewey C N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinformatics, 2011, 12(1): 323. doi: 10.1186/1471-2105-12-323 [16] Pertea M, Pertea G M, Antonescu C M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads[J]. Nature Biotechnology, 2015, 33(3): 290−295. doi: 10.1038/nbt.3122 [17] Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks[J]. Nature Protocols, 2012, 7(3): 562−578. doi: 10.1038/nprot.2012.016 [18] Kang Yujian, Yang Dechang, Kong Lei, et al. CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features[J]. Nucleic Acids Research, 2017, 45(W1): W12−W16. doi: 10.1093/nar/gkx428 [19] Zhao Yingdong, Li M C, Konaté M M, et al. TPM, FPKM, or normalized counts? a comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository[J]. Journal of Translational Medicine, 2021, 19(1): 269. doi: 10.1186/s12967-021-02936-w [20] Wang Likun, Feng Zhixing, Wang Xi, et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data[J]. Bioinformatics, 2010, 26(1): 136−138. doi: 10.1093/bioinformatics/btp612 [21] Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis[J]. BMC Bioinformatics, 2008, 9(1): 559. doi: 10.1186/1471-2105-9-559 [22] Kumar L, Futschik M E. Mfuzz: a software package for soft clustering of microarray data[J]. Bioinformation, 2007, 2(1): 5−7. doi: 10.6026/97320630002005 [23] Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks[J]. Genome Research, 2003, 13(11): 2498−2504. doi: 10.1101/gr.1239303 [24] Chin C H, Chen S H, Wu H H, et al. CytoHubba: identifying hub objects and sub-networks from complex interactome[J]. BMC Systems Biology, 2014, 8(S4): S11. doi: 10.1186/1752-0509-8-S4-S11 [25] Chen Chengjie, Chen Hao, Zhang Yi, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Molecular Plant, 2020, 13(8): 1194−1202. doi: 10.1016/j.molp.2020.06.009 [26] Buchfink B, Xie Chao, Huson D H. Fast and sensitive protein alignment using DIAMOND[J]. Nature Methods, 2015, 12(1): 59−60. doi: 10.1038/nmeth.3176 [27] Koopman P. Sex determination: a tale of two Sox genes[J]. Trends in Genetics, 2005, 21(7): 367−370. doi: 10.1016/j.tig.2005.05.006 [28] Avilion A A, Nicolis S K, Pevny L H, et al. Multipotent cell lineages in early mouse development depend on SOX2 function[J]. Genes & Development, 2003, 17: 126−140. [29] Hong C S, Saint-Jeannet J P. Sox proteins and neural crest development[J]. Seminars in Cell & Developmental Biology, 2005, 16(6): 694−703. [30] Wegner M. All purpose Sox: the many roles of Sox proteins in gene expression[J]. The International Journal of Biochemistry & Cell Biology, 2010, 42(3): 381−390. [31] Wilsker D, Patsialou A, Dallas P B, et al. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development[J]. Cell Growth and Differentiation, 2002, 13(3): 95−106. [32] Cheng Zhongyan, He Tingting, Gao Xiaoming, et al. ZBTB transcription factors: key regulators of the development, differentiation and effector function of T cells[J]. Frontiers in Immunology, 2021, 12: 713294. doi: 10.3389/fimmu.2021.713294 [33] Wasylyk B, Hahn S L, Giovane A. The Ets family of transcription factors[J]. European Journal of Biochemistry, 1993, 211(1/2): 7−18. [34] Wen Qiang, Wang Haitao, Little P J, et al. Forkhead family transcription factor FoxO and neural differentiation[J]. Neurogenetics, 2012, 13(2): 105−113. doi: 10.1007/s10048-012-0320-2 [35] Fatemi M, Wade P A. MBD family proteins: reading the epigenetic code[J]. Journal of Cell Science, 2006, 119(Pt 15): 3033−3037. [36] Paixão-Côrtes V R, Salzano F M, Bortolini M C. Origins and evolvability of the PAX family[J]. Seminars in Cell & Developmental Biology, 2015, 44: 64−74. [37] Wilson V, Conlon F L. The T-box family[J]. Genome Biology, 2002, 3(6): reviews3008. [38] Roussigne M, Kossida S, Lavigne A C, et al. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase[J]. Trends In Biochemical Sciences, 2003, 28(2): 66−69. doi: 10.1016/S0968-0004(02)00013-0 [39] 廖晓婷. 青蛤性腺发育及性别相关基因dmrt1和foxl2的表达研究[D]. 连云港: 江苏海洋大学, 2022.Liao Xiaoting. Study on the gonadal development and expression of sex related genes dmrtl and foxl2 in clam Cyclina sinensis[D]. Lianyungang: Jiangsu Ocean University, 2022. [40] Miller J A, Horvath S, Geschwind D H. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(28): 12698−12703. [41] Voineagu I, Wang Xinchen, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology[J]. Nature, 2011, 474(7351): 380−384. doi: 10.1038/nature10110 [42] Xue Zhigang, Huang K, Cai Chaochao, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing[J]. Nature, 2013, 500(7464): 593−597. doi: 10.1038/nature12364 [43] 高进. 半滑舌鳎(Cynoglossus semilaevis)性腺发育中性别相关基因挖掘及Sox基因家族生物信息学分析[D]. 南京: 南京农业大学, 2016.Gao Jin. Sex related genes mining in the developmental gonad and bioinformatical analysis on sox family of Cynoglossus semilaevis[D]. Nanjing: Nanjing Agricultural University, 2016. [44] 孔宁. 温度、盐度对皱纹盘鲍“97”选群生长发育的影响[D]. 青岛: 中国科学院海洋研究所, 2016.Kong Ning. Effects of temperature and salinity on growth and development of "97" selective breeding population of Haliotis discus hannai Ino[D]. Qingdao: Institute of Oceanology, Chinese Academy of Science, 2016. [45] Bar-Joseph Z. Analyzing time series gene expression data[J]. Bioinformatics, 2004, 20(16): 2493−2503. doi: 10.1093/bioinformatics/bth283 [46] Zalenski A A, Majumder S, De K, et al. An interphase pool of KIF11 localizes at the basal bodies of primary cilia and a reduction in KIF11 expression alters cilia dynamics[J]. Scientific Reports, 2020, 10(1): 13946. doi: 10.1038/s41598-020-70787-4 [47] 庞连慧, 罗双双, 石林林, 等. 团头鲂cdc20基因的序列特征和表达分析[J]. 华中农业大学学报, 2022, 41(4): 226−232. doi: 10.3969/j.issn.1000-2421.2022.4.hznydx202204029Pang Lianhui, Luo Shuangshuang, Shi Linlin, et al. Sequence characteristics and expression analysis of cdc20 gene in Megalobrama amblycephala[J]. Journal of Huazhong Agricultural University, 2022, 41(4): 226−232. doi: 10.3969/j.issn.1000-2421.2022.4.hznydx202204029 [48] Ma Wanying, Du Hua, Kazmi S S U H, et al. UBC gene family and their potential functions on the cellular homeostasis under the elevated pCO2 stress in the diatom Phaeodactylum tricornutum[J]. Ecological Indicators, 2023, 148: 110106. doi: 10.1016/j.ecolind.2023.110106 [49] Borlado L R, Méndez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis[J]. Carcinogenesis, 2008, 29(2): 237−243. doi: 10.1093/carcin/bgm268 [50] Koltowska K, Apitz H, Stamataki D, et al. Ssrp1a controls organogenesis by promoting cell cycle progression and RNA synthesis[J]. Development, 2013, 140(9): 1912−1918. doi: 10.1242/dev.093583 [51] Hou Tingting, Zhang Rufeng, Jian Chongshu, et al. NDUFAB1 confers cardio-protection by enhancing mitochondrial bioenergetics through coordination of respiratory complex and supercomplex assembly[J]. Cell Research, 2019, 29(9): 754−766. doi: 10.1038/s41422-019-0208-x [52] Marsili S, Tichon A, Kundnani D, et al. Gene co-expression analysis of human RNASEH2A reveals functional networks associated with DNA replication, DNA damage response, and cell cycle regulation[J]. Biology(Basel), 2021, 10(3): 221. [53] Skaar J R, Pagano M. Cdh1: a master G0/G1 regulator[J]. Nature Cell Biology, 2008, 10(7): 755−757. doi: 10.1038/ncb0708-755 [54] Zhang Mengna, Li Hui, Guo Mengyu, et al. Vitamin E alleviates pyraclostrobin-induced toxicity in zebrafish(Danio rerio) and its potential mechanisms[J]. Science of The Total Environment, 2024, 922: 171219. doi: 10.1016/j.scitotenv.2024.171219 [55] Lee C Y, Lai Tingyu, Tsai M K, et al. The ubiquitin ligase ZNRF1 promotes caveolin-1 ubiquitination and degradation to modulate inflammation[J]. Nature Communications, 2017, 8(1): 15502. doi: 10.1038/ncomms15502 [56] 刘乔. 基于转录组分析不同生长阶段民猪的特异性表达基因与时序特征[D]. 哈尔滨: 东北农业大学, 2023.Liu Qiao. Analysis of specific expressed genes andtemporal characteristics of Min Pigs at different growth stages based on transcriptomics[D]. Harbin: Northeast Agricultural University, 2023. [57] 赵佳福, 许厚强, 宋书弦, 等. BLM解旋酶基因的克隆、表达载体构建及表达研究[J]. 生物技术通报, 2018, 34(11): 152−159.Zhao Jiafu, Xu Houqiang, Song Shuxian, et al. Cloning and expression vector construction of BLM helicase gene, and its expression analysis[J]. Biotechnology Bulletin, 2018, 34(11): 152−159. [58] Filipovska A, Rackham O. Specialization from synthesis: how ribosome diversity can customize protein function[J]. FEBS Letters, 2013, 587(8): 1189−1197. doi: 10.1016/j.febslet.2013.02.032 [59] Hulm J L, Mcintosh K B, Bonham-Smith P C. Variation in transcript abundance among the four members of the Arabidopsis thaliana RIBOSOMAL PROTEIN S15a gene family[J]. Plant Science, 2005, 169(1): 267−278. doi: 10.1016/j.plantsci.2005.04.001 [60] Latchman D S. Transcription factors: an overview[J]. The International Journal of Biochemistry & Cell Biology, 1997, 29(12): 1305−1312. [61] Yu Jiachen, Zhang Lingling, Li Yangping, et al. Genome-wide identification and expression profiling of the SOX gene family in a bivalve mollusc Patinopecten yessoensis[J]. Gene, 2017, 627: 530−537. doi: 10.1016/j.gene.2017.07.013 [62] 王倩, 郇聘, 刘保忠. 笠贝soxb、mox基因的鉴定及在足原基发育中的表达模式[J]. 海洋与湖沼, 2019, 50(5): 1091−1097. doi: 10.11693/hyhz20190100023Wang Qian, Huan Pin, Liu Baozhong. Expression patterns of soxb and mox genes in Lottia goshimai during the formation of molluscan foot[J]. Oceanologia et Limnologia Sinica, 2019, 50(5): 1091−1097. doi: 10.11693/hyhz20190100023 [63] 梁少帅, 于潇含, 杨丹丹, 等. 栉孔扇贝sox9基因的cDNA克隆及其在不同发育时期性腺中的表达特征[J]. 中国水产科学, 2017, 24(6): 1184−1192. doi: 10.3724/SP.J.1118.2017.17123Liang Shaoshuai, Yu Xiaohan, Yang Dandan, et al. Molecular cloning of sox9 cDNA and its expression characteristies in gonads at different developmental stages of Chlamys farreri[J]. Journal of Fishery Sciences of China, 2017, 24(6): 1184−1192. doi: 10.3724/SP.J.1118.2017.17123 [64] 徐东杰, 谢熙, 王蒙恩, 等. Sox基因家族在水生动物性腺发育中的功能研究进展[J]. 生物学杂志, 2022, 39(3): 97−102. doi: 10.3969/j.issn.2095-1736.2022.03.097Xu Dongjie, Xie Xi, Wang Mengen, et al. Research advance on the function of Sox gene family in aquatic animal gonadal development[J]. Journal of Biology, 2022, 39(3): 97−102. doi: 10.3969/j.issn.2095-1736.2022.03.097 [65] 吴静. SOX2在猪早期胚胎发育中生物学功能及Hippo信号通路对其表达调控的研究[D]. 哈尔滨: 东北农业大学, 2023.Wu Jing. Study on the biological function of SOX2 and the regulation of its expression by the hippo signaling pathway in porcineearly embryonic development[D]. Harbin: Northeast Agricultural University, 2023. -





 下载:
下载: