Study on denitrification process of sediment in the Liaohe EstuaryAnalysis of the abundance of denitrification functional genes and the community structure of nirK-type bacteria
-
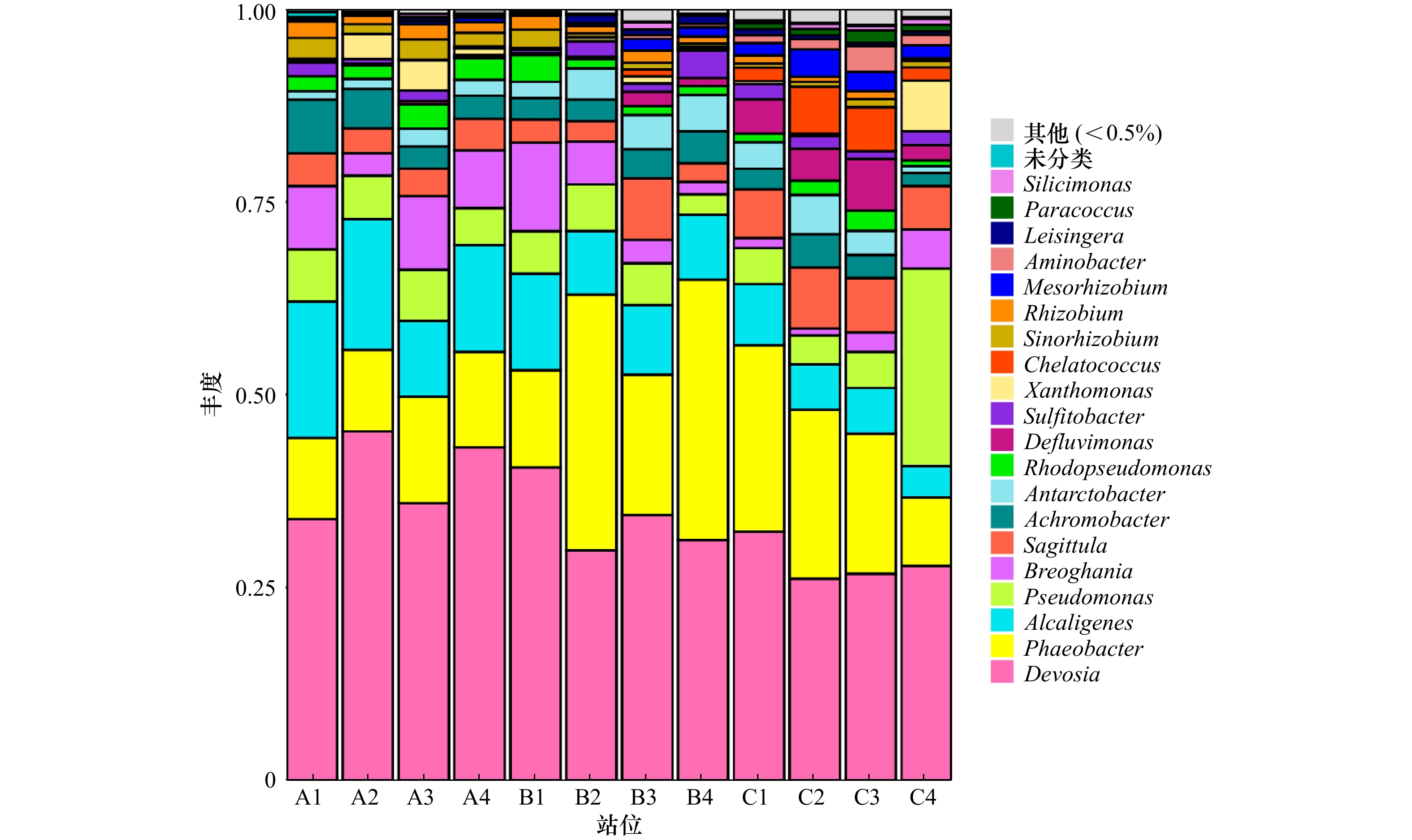
摘要: 近年来,辽河口沿岸人类活动频繁,工农业活动发达,大量的氮、磷等营养物质输入至感潮河段继而排放入海,导致辽河口水体富营养化程度加剧。沉积物区域是反硝化作用的重要发生地和微生物富集地。微生物能够将沉积物中的硝酸盐及亚硝酸盐还原为N2O和N2释放到大气中,进而减轻河口生态系统氮负荷。本文中的实验采用定量PCR技术测定辽河口表层沉积物反硝化过程功能基因narG、nirK、norB、nosZ的基因丰度,结果表明,主导硝酸盐还原的narG功能基因丰度最高。使用高通量测序技术对nirK型功能基因进行测序,结果显示,在辽河口反硝化细菌中Devosia、Phaeobacter、Alcaligenes、Pseudomonas菌属丰度较高。反硝化功能基因的丰度主要受到沉积物粒径的影响,norB基因丰度与多个环境因子显著相关。nirK型反硝化细菌的群落结构和多样性主要受盐度、pH、溶解氧以及NO2−的影响。研究分析了反硝化功能基因丰度和细菌群落结构及其主要影响因子,其结果为辽河口水体富营养化中氮元素归趋的认知提供理论依据。Abstract: In recent years, the water quality of the Liaohe Estuary has been declining, and a large input of nitrogen has increased the burden on the environment. The denitrification process releases nitrogen from sediments into the atmosphere in the form of N2O and N2, which reduces the nitrogen load in the estuarine ecosystem. In this study, quantitative PCR was used to determine the levels of four denitrification functional genes narG, nirK, norB and nosZ present in the surface sediments of the Liaohe Estuary. The narG gene, which dominates nitrate reduction, was found to be the most abundant. The nirK-type denitrification functional gene was sequenced by high-throughput sequencing. The results showed that the genus of denitrifying bacteria in the Liaohe Estuary was mainly Devosia, Phaeobacter, Alcaligenes, Pseudomonas. The abundance of denitrification function genes is mainly affected by sediment properties, except that norB gene is significantly related to many environmental factors. The community structure and diversity of nirK type denitrifying bacteria are mainly affected by salinity, pH, dissolved oxygen and
${\rm {NO}}_2^- $ contents. The effects of environmental factors on the abundance and community structure of denitrifying bacteria were studied. The results provided a theoretical basis for alleviating the fate of nitrogen elements in the eutrophication of Liaohe Estuary. -
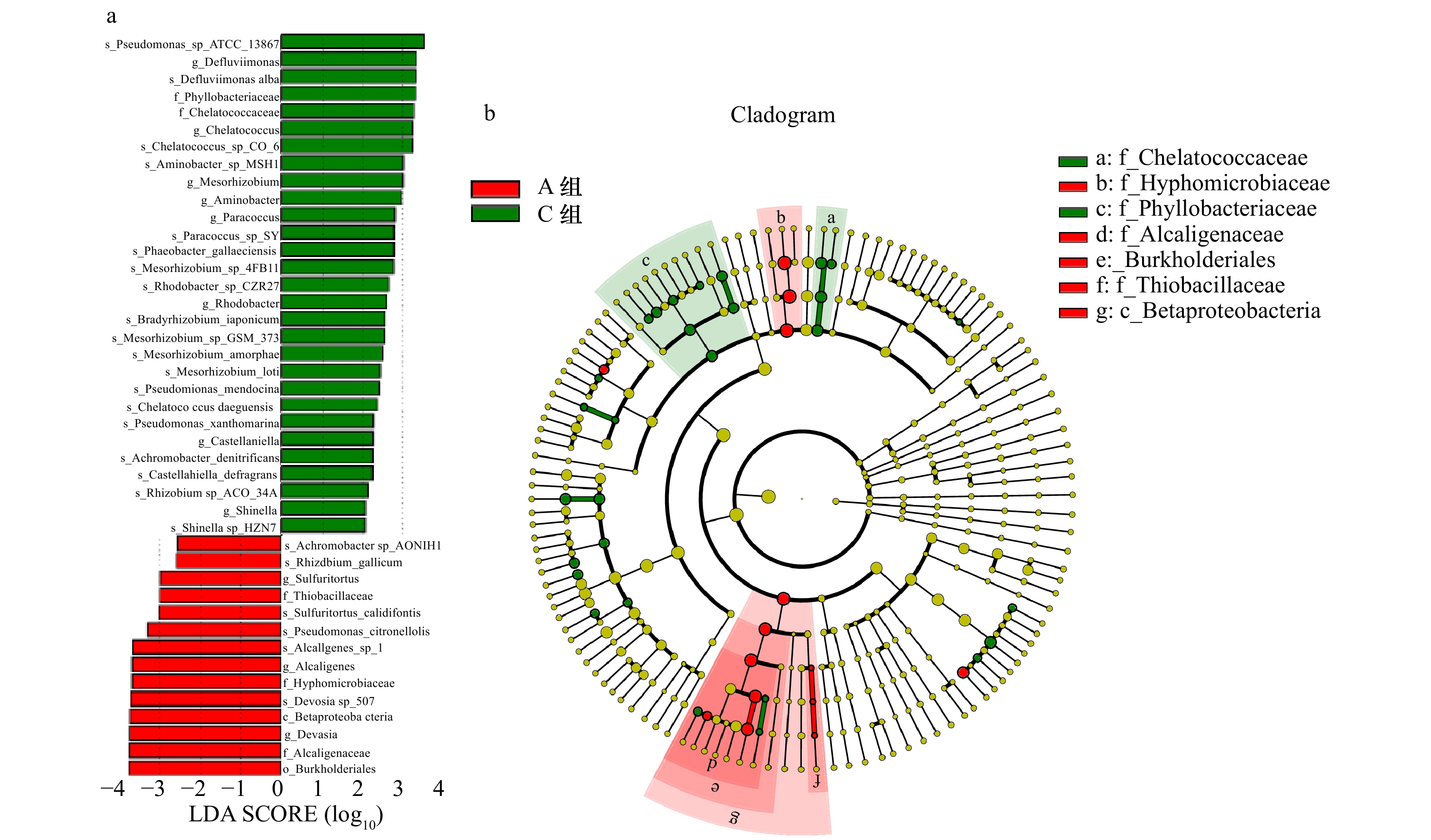
图 6 基于LEfSe分析的辽河口沉积物中nirK型反硝化细菌的丰度柱状图(a)和进化分支图(b)
LDA SCORE是指用线性判别分析(LDA)对数据进行降维和评估差异显著的物种的影响力
Fig. 6 Abundance histogram (a) and evolutionary branch map (b) of nir K-type denitrifying bacteria in the sediments of the Liaohe Estuary based on LEfSe analysis
LDA SCORE means using linear discriminant analysis (LDA) to reduce the dimension of data and evaluate the influence of species with significant differences
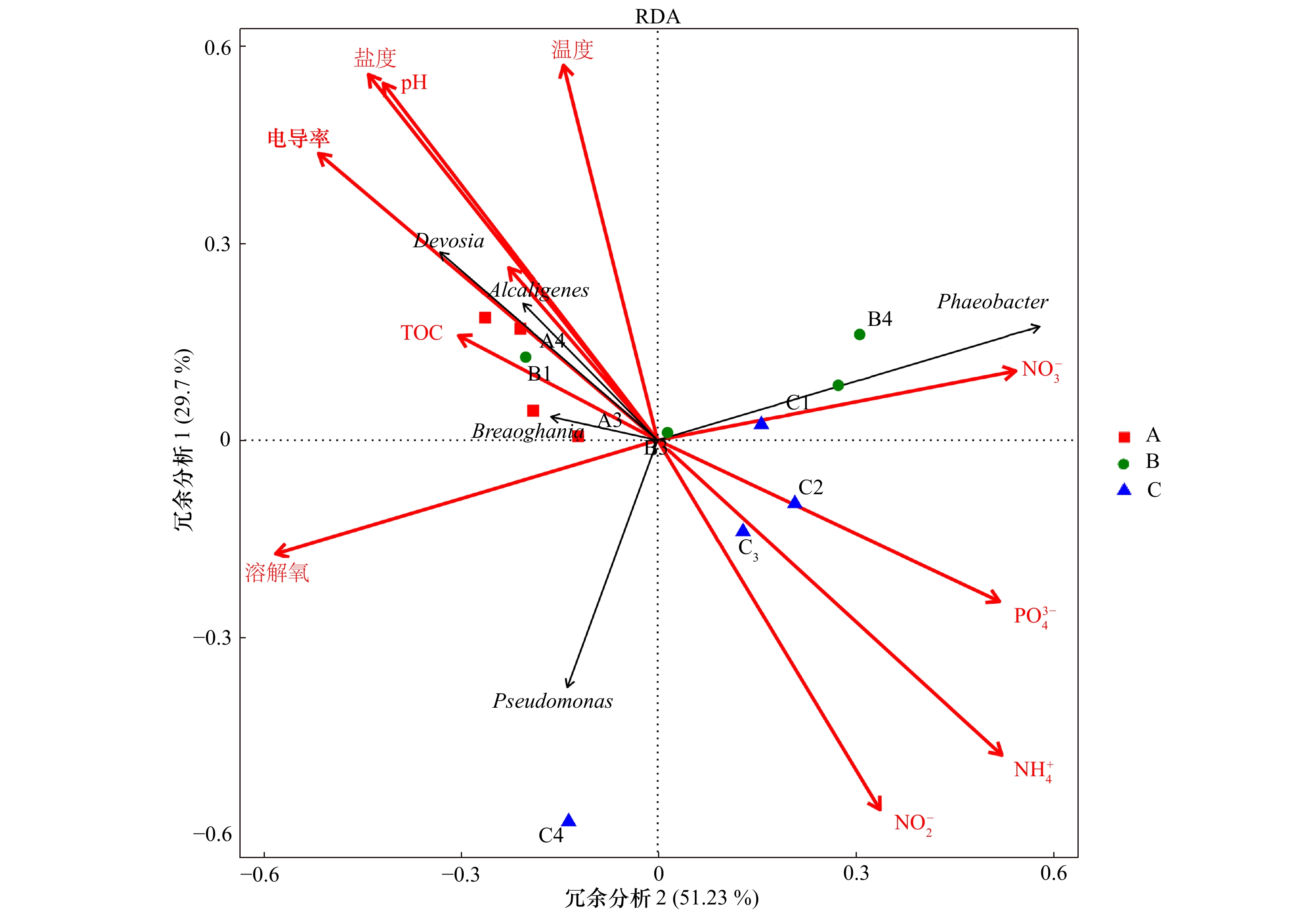
图 7 nirK型反硝化细菌群落与环境因子变量的冗余分析图
横轴和纵轴分别在两个主要维度上解释了微生物群落组成中携带nirK基因的突变量
Fig. 7 Redundancy analysis of nirK-type denitrifying bacteria community and environmental factor variables
The horizontal axils and the vertical axils explained the variation of nirK gene in microbial community composition in two main dimensions
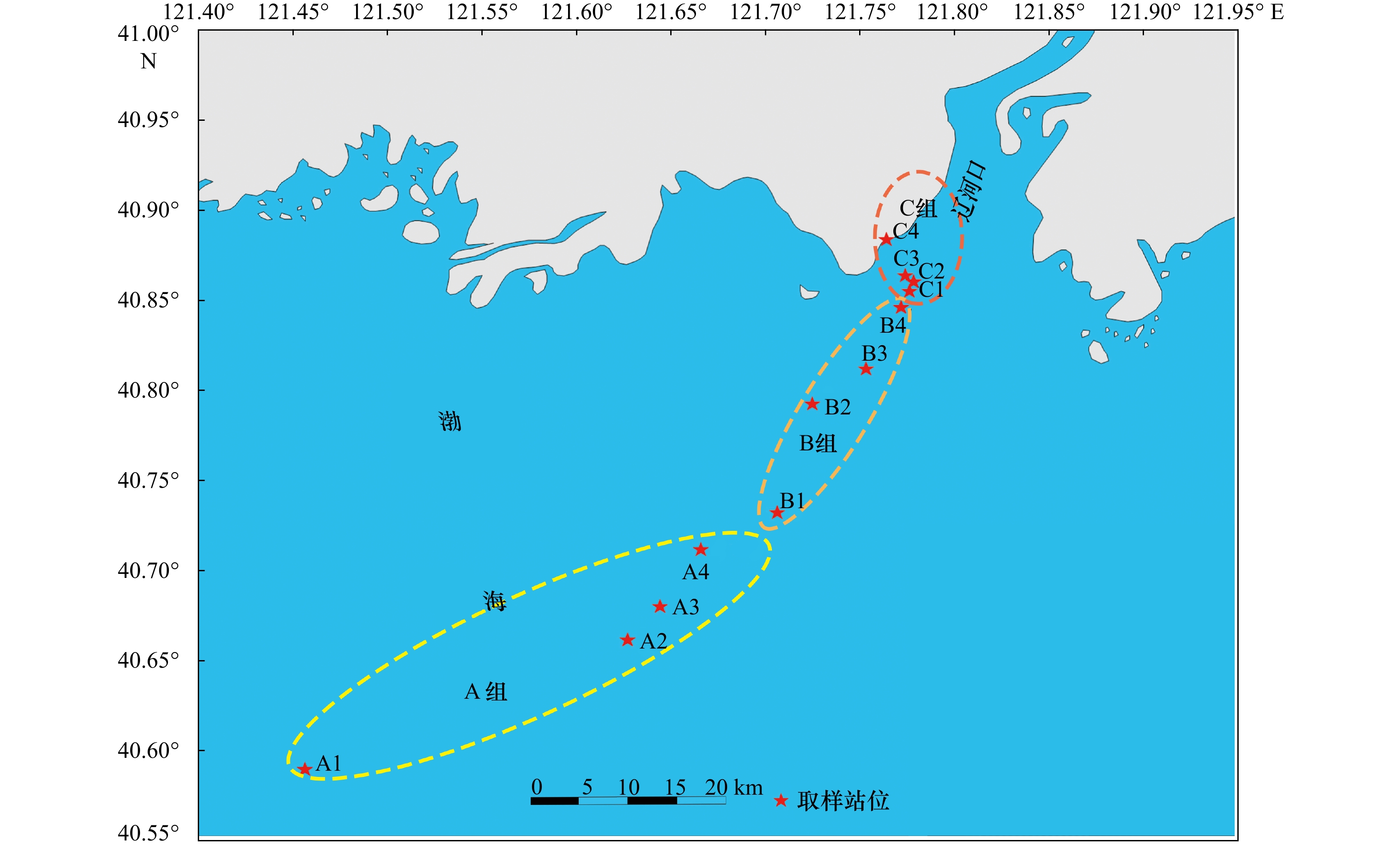
表 1 辽河口底层水中环境因子理化参数[11]
Tab. 1 Physical and chemical parameters of environmental factors in bottom water of the Liaohe Estuary[11]
站位 温度/℃ 盐度 电导率/mS·cm−1 pH DO浓度/mg·L−1 $ {{\rm {NH}}_4^+} $浓度/μmol·L−1 $ {{\rm {NO}}_2^-} $浓度/μmol·L−1 $ {{\rm {NO}}_3^- }$浓度/μmol·L−1 $ {{\rm {PO}}_4^{3-}} $浓度/μmol·L−1 A1 26.2 31.5 47.3 8.00 6.3 1.08 0.24 0.80 0.11 A2 27.0 28.7 44.6 7.99 6.3 1.11 0.32 7.27 0.13 A3 26.9 27.7 43.2 7.98 6.4 1.18 0.39 8.95 0.18 A4 26.7 25.5 40.0 7.94 6.1 1.49 0.48 12.32 0.01 B1 26.5 19.6 31.5 7.86 6.3 5.89 0.37 17.02 0.29 B2 27.0 20.7 33.1 7.92 6.1 5.61 0.35 15.27 0.20 B3 26.8 19.8 32.0 7.88 5.0 5.94 0.40 15.95 0.28 B4 26.5 13.1 21.8 7.72 3.3 9.50 0.61 15.23 0.27 C1 25.9 7.0 13.3 7.62 4.0 9.22 0.60 18.63 0.34 C2 26.0 5.5 9.7 7.57 3.1 14.28 1.02 22.06 0.40 C3 25.9 2.4 4.5 7.54 6.2 13.61 0.94 21.98 0.31 C4 25.7 0.8 15.8 7.54 6.9 11.83 0.93 5.08 0.23 表 2 辽河口表层沉积物中环境因子理化参数[11]
Tab. 2 Physical and chemical parameters of environmental factors in surface sediments of the Liaohe Estuary[11]
站位 TOC含量/% TP浓度/μg·g−1 TN浓度/μg·g−1 黏土含量/% 粉砂含量/% 砂含量/% A1 0.82 185.0 34.7 31.18 58.48 10.34 A2 0.43 57.2 40.2 23.53 51.55 24.92 A3 0.53 94.4 65.3 26.24 61.35 12.41 A4 0.56 90.3 60.5 31.01 60.13 8.85 B1 0.49 97.1 131.0 37.00 62.97 0.03 B2 0.70 99.6 298.0 14.57 56.67 28.76 B3 0.60 102.0 136.0 21.46 61.57 16.97 B4 0.14 117.0 76.5 12.60 59.54 27.87 C1 0.34 109.0 87.9 18.01 66.96 15.04 C2 0.34 47.8 38.3 4.64 47.36 47.99 C3 0.46 84.4 90.5 18.47 53.82 27.71 C4 0.34 26.6 30.1 2.67 12.45 84.88 表 3 反硝化功能基因的引物
Tab. 3 Primers for denitrification genes
基因 引物 5′-3′ 序列 参考文献 nirK FlaCu ATC ATG GTS CTG CCG CG [12] R3Cu GCC TCG ATC AGR TTG TGG TT nosZ nosZ1126F GGG CTB GGG CCR TTG CA [13] nosZ1138R GAA GCG RTC CTT SGA RAA CTTG narG narG571F CCG ATY CCG GCV ATG TCS AT [13] narG773R GGN ACG TTN GAD CCC CA norB norB1F CGN GAR TTY CTS GAR CAR CC [14] norB3R CCY TCV ACC CAG ASA TGC AC 表 4 辽河口nirK型反硝化细菌丰度及多样性
Tab. 4 Abundance and diversity of nirK
-type denitrifying bacteria in the Liaohe Estuary 采样点 OTUs
个数有效序
列数Coverage
/%Chao1
指数ACE
指数Shannon
指数Simpson
指数A1 112 37071 99.96 138.25 127.22 2.66 0.146 A2 96 36947 99.99 97.00 97.99 2.28 0.230 A3 121 36164 99.99 121.27 122.16 2.71 0.151 A4 132 30487 99.95 140.67 139.96 2.54 0.201 B1 126 30315 99.94 146.00 135.69 2.52 0.188 B2 136 32816 99.97 138.80 139.67 2.59 0.169 B3 237 37310 99.96 244.09 242.59 3.25 0.122 B4 162 31673 99.94 183.86 172.69 2.78 0.144 C1 236 37745 99.97 245.43 240.57 3.32 0.103 C2 305 37274 99.94 319.62 313.62 3.62 0.077 C3 291 36136 99.97 293.77 293.63 3.60 0.088 C4 160 33334 99.89 248.00 267.54 2.82 0.140 -
[1] Smith D P, Thrash J C, Nicora C D, et al. Proteomic and transcriptomic analyses of “Candidatus Pelagibacter ubique”describe the first PII-independent response to nitrogen limitation in a free-living Alphaproteobacterium[J]. mBio, 2013, 4(6): e00133−12. [2] Herbert R A. Nitrogen cycling in coastal marine ecosystems[J]. FEMS Microbiology Reviews, 1999, 23(5): 563−590. doi: 10.1111/j.1574-6976.1999.tb00414.x [3] Helen D, Kim H, Tytgat B, et al. Highly diverse nirK genes comprise two major clades that harbour ammonium-producing denitrifiers[J]. BMC Genomics, 2016, 17(1): 155. doi: 10.1186/s12864-016-2465-0 [4] Zumft W G. Cell biology and molecular basis of denitrification[J]. Microbiology and Molecular Biology Reviews, 1997, 61(4): 533−616. doi: 10.1128/.61.4.533-616.1997 [5] Penton C R, Johnson T A, Quensen J F, et al. Functional genes to assess nitrogen cycling and aromatic hydrocarbon degradation: primers and processing matter[J]. Frontiers in Microbiology, 2013, 4: 279. [6] Santoro A E, Boehm A B, Francis C A. Denitrifier community composition along a nitrate and salinity gradient in a coastal aquifer[J]. Applied and Environmental Microbiology, 2006, 72(3): 2102−2109. doi: 10.1128/AEM.72.3.2102-2109.2006 [7] Jones C M, Hallin S. Ecological and evolutionary factors underlying global and local assembly of denitrifier communities[J]. The ISME Journal, 2010, 4(5): 633−641. doi: 10.1038/ismej.2009.152 [8] 陶怡乐, 温东辉. 细菌硝酸盐异化还原成铵过程及其在河口生态系统中的潜在地位与影响[J]. 微生物学通报, 2016, 43(1): 172−181.Tao Yile, Wen Donghui. Dissimilatory nitrate reduction to ammonium: The potential and impacts in estuarine regions[J]. Microbiology China, 2016, 43(1): 172−181. [9] 袁晓敏, 杨继松, 刘凯, 等. 盐分对辽河口湿地土壤DOC及CO2生成的影响[J]. 生态学杂志, 2017, 36(8): 2111−2117.Yuan Xiaomin, Yang Jisong, Liu Kai, et al. Effects of salinity on DOC concentration and CO2 production of wetland soil in Liaohe Estuarine[J]. Chinese Journal of Ecology, 2017, 36(8): 2111−2117. [10] Bu Hongmei, Meng Wei, Zhang Yuan. Nitrogen pollution and source identification in the Haicheng River basin in Northeast China[J]. Science of the Total Environment, 2011, 409(18): 3394−3402. doi: 10.1016/j.scitotenv.2011.05.030 [11] 张慧珍, 常永凯, 陈泉睿, 等. 辽河口沉积物中古菌和细菌群落结构分析[J]. 海洋学报, 2018, 40(6): 113−130.Zhang Huizhen, Chang Yongkai, Chen Quanrui, et al. Community structure analysis of archaea and bacteria in sediments of Liaohe Estuary[J]. Haiyang Xuebao, 2018, 40(6): 113−130. [12] Hallin S, Lindgren P E. PCR detection of genes encoding nitrite reductase in denitrifying bacteria[J]. Applied and Environmental Microbiology, 1999, 65(4): 1652−1657. doi: 10.1128/AEM.65.4.1652-1657.1999 [13] Chen Zhe, Liu Jinbo, Wu Minna, et al. Differentiated response of denitrifying communities to fertilization regime in paddy soil[J]. Microbial Ecology, 2012, 63(2): 446−459. doi: 10.1007/s00248-011-9909-5 [14] Casciotti K L, Ward B B. Phylogenetic analysis of nitric oxide reductase gene homologues from aerobic ammonia-oxidizing bacteria[J]. FEMS Microbiology Ecology, 2005, 52(2): 197−205. doi: 10.1016/j.femsec.2004.11.002 [15] Dang Hongyue, Li Jing, Chen Ruipeng, et al. Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China[J]. Applied and Environmental Microbiology, 2010, 76(14): 4691−4702. doi: 10.1128/AEM.02563-09 [16] Jiang Xiaoliang, Yao Lu, Guo Laodong, et al. Multi-scale factors affecting composition, diversity, and abundance of sediment denitrifying microorganisms in Yangtze lakes[J]. Applied Microbiology and Biotechnology, 2017, 101(21): 8015−8027. doi: 10.1007/s00253-017-8537-5 [17] Yang Jisong, Zhan Chao, Li Yunzhao, et al. Effect of salinity on soil respiration in relation to dissolved organic carbon and microbial characteristics of a wetland in the Liaohe River estuary, Northeast China[J]. Science of the Total Environment, 2018, 642: 946−953. doi: 10.1016/j.scitotenv.2018.06.121 [18] Mahjoubi M, Aliyu H, Cappello S, et al. The genome of Alcaligenes aquatilis strain BU33N: insights into hydrocarbon degradation capacity[J]. PLoS One, 2019, 14(9): e0221574. doi: 10.1371/journal.pone.0221574 [19] Jiang Yan, Wen Jianping, Bai Jing, et al. Biodegradation of phenol at high initial concentration by Alcaligenes faecalis[J]. Journal of Hazardous Materials, 2007, 147(1/2): 672−676. [20] Chen Yong, Zhu Sidong, Lin Danqiu, et al. Devosia Naphthalenivorans sp. nov., isolated from East Pacific Ocean sediment[J]. International Journal of Systematic and Evolutionary Microbiology, 2019, 69(7): 1974−1979. doi: 10.1099/ijsem.0.003410 [21] Park S, Jung Y T, Kim S, et al. Devosia confluentis sp. nov., isolated from the junction between the ocean and a freshwater lake, and reclassification of two Vasilyevaea species as Devosia enhydra comb. nov. and Devosia mishustinii comb. nov[J]. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(10): 3935−3941. doi: 10.1099/ijsem.0.001291 [22] Wolińska A, Kuźniar A, Zielenkiewicz U, et al. Metagenomic analysis of some potential nitrogen-fixing bacteria in arable soils at different formation processes[J]. Microbial Ecology, 2017, 73(1): 162−176. doi: 10.1007/s00248-016-0837-2 [23] Rivas R, Willems A, Subba-Rao N S, et al. Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India[J]. Systematic and Applied Microbiology, 2003, 26(1): 47−53. doi: 10.1078/072320203322337308 [24] Fu Yingnan, Wang Rui, Zhang Zilian, et al. Complete genome sequence of the d-Amino acid catabolism bacterium Phaeobacter sp. Strain JL2886, isolated from deep seawater of the South China Sea[J]. Genome Announcements, 2016, 4(5): e00913−16. [25] Gram L, Rasmussen B B, Wemheuer B, et al. Phaeobacter inhibens from the Roseobacter clade has an environmental niche as a surface colonizer in harbors[J]. Systematic and Applied Microbiology, 2015, 38(7): 483−493. doi: 10.1016/j.syapm.2015.07.006 [26] Trautwein K, Hensler M, Wiegmann K, et al. The marine bacterium Phaeobacter inhibens secures external ammonium by rapid buildup of intracellular nitrogen stocks[J]. FEMS Microbiology Ecology, 2018, 94(10): 1−14. [27] Perkins T L, Clements K, Baas J H, et al. Sediment composition influences spatial variation in the abundance of human pathogen indicator bacteria within an estuarine environment[J]. PLoS One, 2014, 9(11): e112951. doi: 10.1371/journal.pone.0112951 [28] Luo Ling, Zhou Zhichao, Gu Jidong. Distribution, diversity and abundance of bacterial laccase-like genes in different particle size fractions of sediments in a subtropical mangrove ecosystem[J]. Ecotoxicology, 2015, 24(7/8): 1508−1516. [29] Zaghmouri I, Michotey V D, Armougom F, et al. Salinity shifts in marine sediment: importance of number of fluctuation rather than their intensities on bacterial denitrifying community[J]. Marine Pollution Bulletin, 2018, 130: 76−83. doi: 10.1016/j.marpolbul.2018.03.020 [30] Liu Wenzhi, Yao Lu, Jiang Xiaoliang, et al. Sediment denitrification in Yangtze lakes is mainly influenced by environmental conditions but not biological communities[J]. Science of the Total Environment, 2018, 616−617: 978−987. doi: 10.1016/j.scitotenv.2017.10.221 [31] Zhou Shilei, Huang Tinglin, Zhang Chunhua, et al. Illumina MiSeq sequencing reveals the community composition of NirS-Type and NirK-Type denitrifiers in Zhoucun reservoir—a large shallow eutrophic reservoir in northern China[J]. RSC Advances, 2016, 6(94): 91517−91528. doi: 10.1039/C6RA18017E [32] Fan Haoxin, Bolhuis H, Stal L J. Denitrification and the denitrifier community in coastal microbial mats[J]. FEMS Microbiology Ecology, 2015, 91(3): 1−11. -





 下载:
下载: