Comparison of the pyloric caecum microbiota community structure between the wild and farmed Larimichthys crocea in Sansha Bay
-
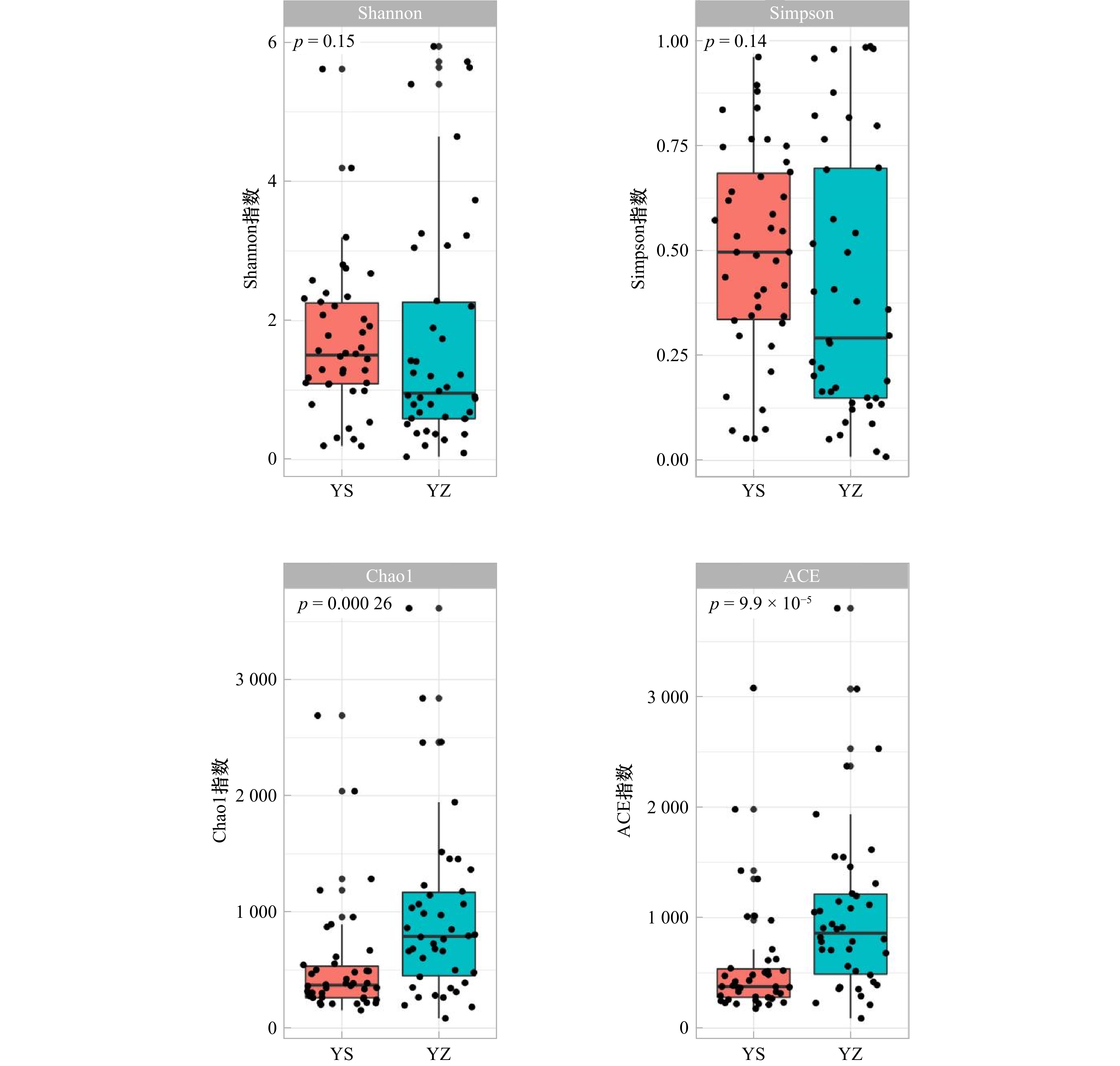
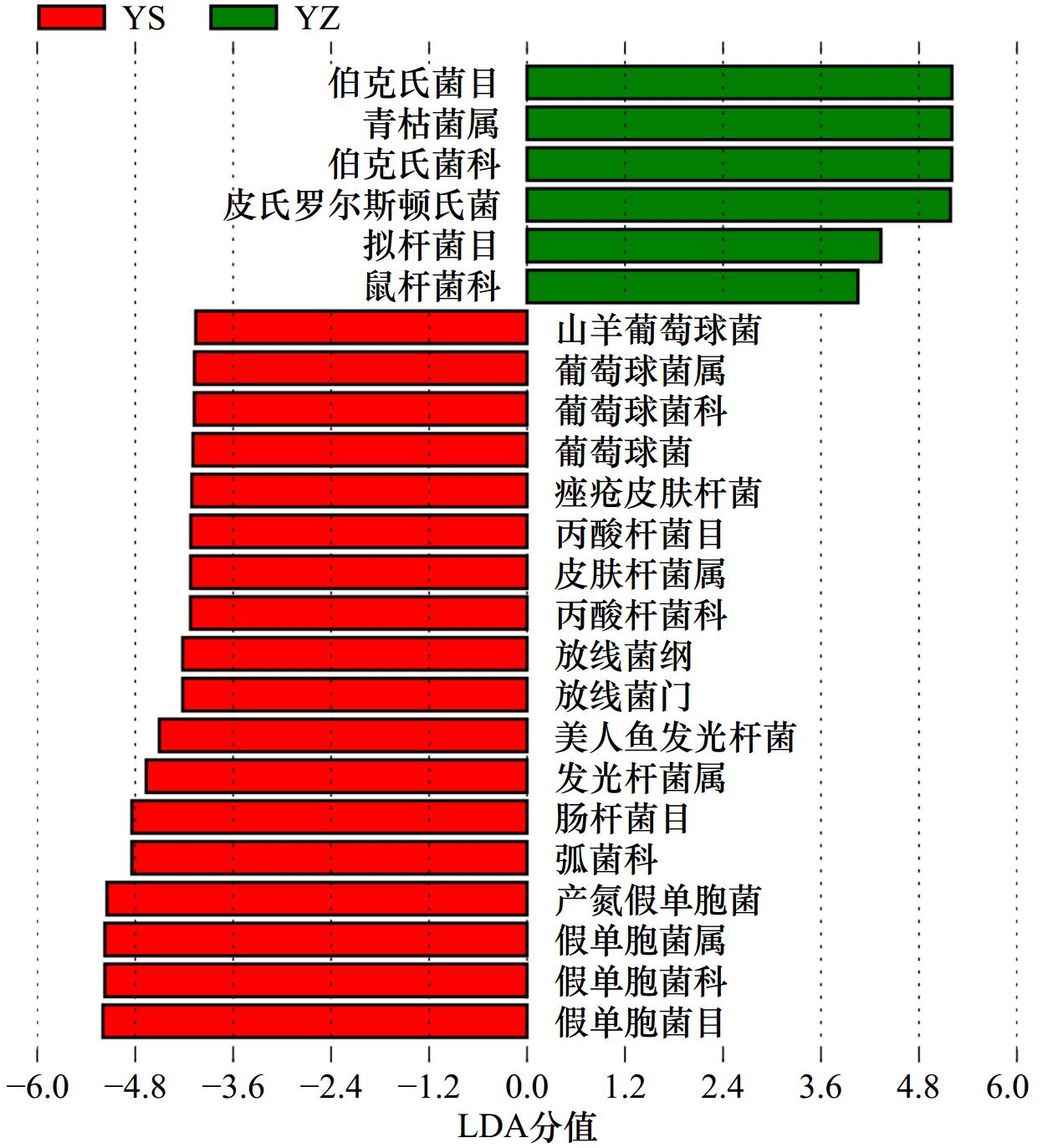
摘要: 大黄鱼(Larimichthys crocea)幽门盲囊呈半封闭结构,保留了部分早期孵化阶段的微生物特征,是用于野生和养殖群体溯源的理想部位。本研究采用高通量测序技术,对三沙湾大黄鱼野生和养殖群体的幽门盲囊微生物的α多样性、核心菌群相对丰度、网络关系等进行分析,构建随机森林模型进行群体溯源分析。结果显示,养殖群体的幽门盲囊微生物有着更多特有操作分类单元(Operational Taxonomic Unit, OTU),其α多样性显著高于野生群体。幽门盲囊微生物的优势菌群有变形菌门(Proteobacteria)、厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidota)、放线菌门(Actinobacteria)及酸杆菌门(Acidobacteria),且各菌群在野生和养殖群体中的相对丰度差异显著。微生物网络分析结果显示,野生和养殖群体的幽门盲囊微生物群落结构差异明显,野生群体拥有更高的负边缘/正边缘比率和模块性以及更少的节点数和连接数。基于此,我们构建了随机森林分类群体溯源预测模型,其准确率(Accuracy, ACC)达92.31%,Kappa系数为0.845 2,ROC曲线下面积为0.952 4,利用此模型识别三沙湾大黄鱼野生和养殖群体的准确率分别可达91.67%和92.86%。综上所述,三沙湾大黄鱼野生和养殖群体的幽门盲囊微生物群落结构差异显著,利用幽门盲囊微生物追踪鱼类来源的方法是可行且有效的,研究结果为区分三沙湾大黄鱼野生和养殖群体提供了新思路。Abstract: The semienclosed pyloric caecum of Larimichthys crocea is an ideal organ to perform the host source tracking, as it containes several local bacteria colonized during early development. The alpha diversity, relative abundance of core bacteria and network relationship of the pyloric caecum microbiota in L. crocea from Sansha Bay were analyzed using the Illumina high-throughput sequencing. Furthermore, the random forest model was used to predict the population source (the wild population or the farmed population). The results showed that the farmed fish had more unique OUT and higher alpha diversity than the wild one. The wild and farmed fish significantly differed in the relative abundance of dominant bacteria (Proteobacteria, Firmicutes, Bacteroidota, Actinobacteria and Acidobacteria) (p < 0.05). The result of network analysis showed that the wild fish had higher ratio of negative to positive edges and modularity, but fewer nodes and edges than the farmed fish. Furthermore, a random forest classification prediction model with the accuracy of 92.31%, the Kappa coefficient of 0.8452, and an area under the ROC curve of 0.952 4 was constructed. Using this prediction model, the accuracy of source identification for the wild and farmed fish was 91.67% and 92.86%, respectively. Overall, the structure of microbial communities in the pyloric caecum of L. crocea was different between the wild and farmed populations, making it a potential marker for host source tracking. Our findings provide new insights into distinguishing wild and farmed L. crocea.
-
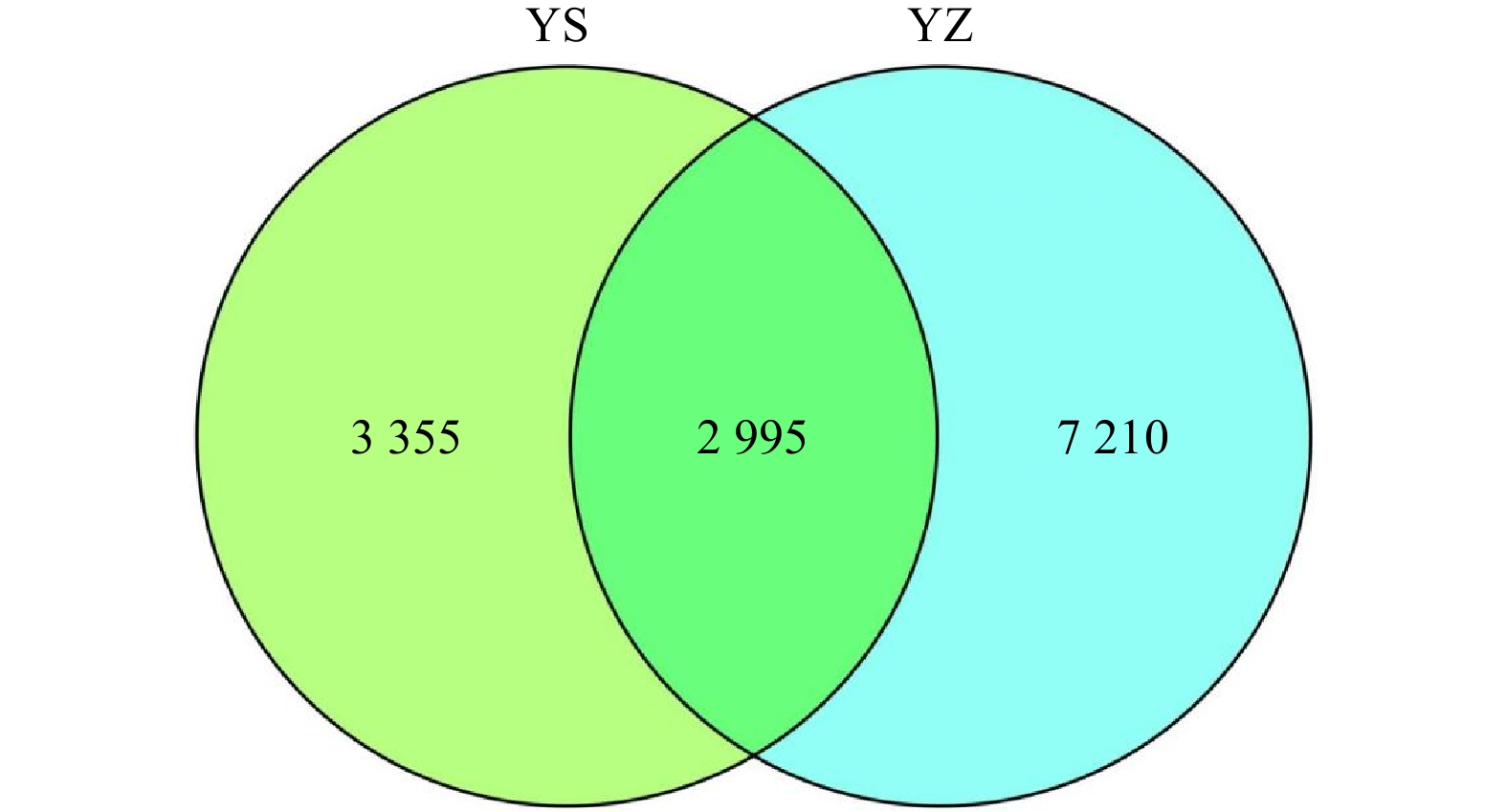
图 1 三沙湾大黄鱼野生群体(YS)和养殖群体(YZ)幽门盲囊微生物的Venn图
不同颜色代表不同组样本,图中数字表示OTU数量
Fig. 1 Venn diagram of the pyloric caecum microbiota of the wild population (YS) and farmed population (YZ) of Larimichthys crocea in Sansha Bay
Different colors indicate different groups, and the numbers within cycles indicate the number of OTU
图 7 三沙湾大黄鱼野生群体(YS)和养殖群体(YZ)幽门盲囊微生物的网络关系
图中点的不同颜色代表不同门,点的大小表示连接到该节点边缘数的多少;各点之间相互连线的粗细表示相关性系数绝对值的大小(p < 0.05, r > 0.6)
Fig. 7 The co-occurrence networks of the pyloric caecum microbiota of the wild population (YS) and farmed population (YZ) of Larimichthys crocea in Sansha Bay
Different colors of nodes indicate different bacterial phyla, and the size of nodes indicates the number of connected edges; the thickness of the connecting line between each point indicates the absolute value of correlation coefficient (p < 0.05, r > 0.6)
表 1 三沙湾大黄鱼野生群体(YS)和养殖群体(YZ)幽门盲囊微生物的网络图特征参数
Tab. 1 Characteristic parameters of network diagram of the pyloric caecum microbiota of the wild population (YS) and farmed population (YZ) of Larimichthys crocea in Sansha Bay
网络图特征参数 YS YZ 节点数 163 306 连接数 1 561 12 309 正连接数 1 525 12 125 负连接数 36 184 负/正连接数比 0.023 0.015 平均度 5.423 52.118 平均聚集系数 0.461 0.631 模块性 0.695 0.292 表 2 随机森林模型参数
Tab. 2 The parameters of the random forest model
模型参数 数值 准确率 92.31% Kappa系数 0.8452 敏感性 92.86% 特异性 91.67% 正预测值 92.86% 负预测值 91.67% 平衡精度 92.26% ROC 曲线下面积 0.952 4 表 3 测试集中随机森林预测模型分类结果
Tab. 3 Classification results of the test set using the random forest prediction model
预测分类 实际分类 准确率/% YS YZ YS 11 1 91.67 YZ 1 13 92.86 -
[1] 阮成旭, 袁重桂, 陶翠丽, 等. 不同养殖模式对大黄鱼肉质的影响[J]. 水产科学, 2017, 36(5): 623−627.Ruan Chengxu, Yuan Chonggui, Tao Cuili, et al. Influence of culture patterns on flesh quality of large yellow croaker Pseudosciaena crocea[J]. Fisheries Science, 2017, 36(5): 623−627. [2] 胡兵. 大黄鱼系列配合饲料的应用现状[J]. 中国水产, 2015(3): 48−50.Hu Bing. The application status of large yellow croaker series formula feed[J]. China Fisheries, 2015(3): 48−50. [3] 黄伟强, 纪炜炜, 付婧, 等. 三沙湾大黄鱼网箱养殖衍生有机物的沉降特征[J]. 中国水产科学, 2020, 27(6): 709−719.Huang Weiqiang, Ji Weiwei, Fu Jing, et al. Sedimentation characteristics of aquaculture-derived organic matter from a large yellow croaker ( Larimichthys crocea) cage farm in Sansha Bay[J]. Journal of Fishery Sciences of China, 2020, 27(6): 709−719. [4] 王萱, 刘义峰, 郭伟. 近十年三沙湾海水增养殖区环境质量状况与变化趋势评价[J]. 渔业研究, 2019, 41(6): 519−525.Wang Xuan, Liu Yifeng, Guo Wei. Evaluation of environmental quality and change trend in Sansha Bay mariculture area in recent ten years[J]. Journal of Fisheries Research, 2019, 41(6): 519−525. [5] 王映, 柯巧珍, 刘家富, 等. 大黄鱼养殖群体和野生群体形态、鳞片及耳石特征比较[J]. 海洋渔业, 2016, 38(2): 149−156.Wang Ying, Ke Qiaozhen, Liu Jiafu, et al. Comparison on morphology, scales and otolith characteristics between cultured stock and wild stock of Larimichthys crocea[J]. Marine Fisheries, 2016, 38(2): 149−156. [6] 张其永, 洪万树. 官井洋大黄鱼资源的兴衰演变及其修复对策[J]. 海洋渔业, 2015, 37(2): 179−186.Zhang Qiyong, Hong Wanshu. Resource status and remediation strategy for large yellow croaker in Guanjingyang Bay[J]. Marine Fisheries, 2015, 37(2): 179−186. [7] 丁爱侠, 贺依尔. 岱衢族大黄鱼放流增殖试验[J]. 南方水产科学, 2011, 7(1): 73−77.Ding Aixia, He Yi’er. Test on release and proliferation of Pseudosciaena crocea in Daiquyang sea area[J]. South China Fisheries Science, 2011, 7(1): 73−77. [8] 张其永, 洪万树, 杨圣云, 等. 大黄鱼增殖放流的回顾与展望[J]. 现代渔业信息, 2010, 25(12): 3−5, 12.Zhang Qiyong, Hong Wanshu, Yang Shengyun, et al. Review and prospects in the restocking of the large yellow croaker ( Larimichthys crocea)[J]. Modern Fisheries Information, 2010, 25(12): 3−5, 12. [9] 农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 中国渔业统计年鉴2022[M]. 北京: 中国农业出版社, 2022.Ministry of Agriculture and Rural Affairs of the People’s Republic of China, National Fisheries Technology Extension Center, China Society of Fisheries. China Fisheries Statistical Yearbook 2022[M]. Beijing: China Agriculture Press, 2022. [10] 郭全友, 邢晓亮, 姜朝军, 等. 野生和养殖大黄鱼( Larimichthys crocea)品质特征与差异性探究[J]. 现代食品科技, 2019, 35(10): 92−101.Guo Quanyou, Xing Xiaoliang, Jiang Chaojun, et al. Quality characteristics and differences of wild and cultured large yellow croakers ( Larimichthys crocea)[J]. Modern Food Science and Technology, 2019, 35(10): 92−101. [11] 孟晓林, 李文均, 聂国兴. 鱼类肠道菌群影响因子研究进展[J]. 水产学报, 2019, 43(1): 143−155.Meng Xiaolin, Li Wenjun, Nie Guoxing. Effect of different factors on the fish intestinal microbiota[J]. Journal of Fisheries of China, 2019, 43(1): 143−155. [12] Clements K D, Angert E R, Montgomery W L, et al. Intestinal microbiota in fishes: what’s known and what’s not[J]. Molecular Ecology, 2014, 23(8): 1891−1898. doi: 10.1111/mec.12699 [13] Egerton S, Culloty S, Whooley J, et al. The gut microbiota of marine fish[J]. Frontiers in Microbiology, 2018, 9: 873. doi: 10.3389/fmicb.2018.00873 [14] Ganguly S, Prasad A. Microflora in fish digestive tract plays significant role in digestion and metabolism[J]. Reviews in Fish Biology and Fisheries, 2012, 22(1): 11−16. doi: 10.1007/s11160-011-9214-x [15] Wang A R, Ran C, Ringø E, et al. Progress in fish gastrointestinal microbiota research[J]. Reviews in Aquaculture, 2018, 10(3): 626−640. doi: 10.1111/raq.12191 [16] 何娇娇, 王萍, 冯建, 等. 玉米蛋白粉对大黄鱼生长、肠道组织结构及肠道菌群的影响[J]. 中国水产科学, 2018, 25(2): 361−372. doi: 10.3724/SP.J.1118.2018.17180He Jiaojiao, Wang Ping, Feng Jian, et al. Effects of replacing fish meal with corn gluten meal on growth, intestinal histology, and intestinal microbiota of large yellow croaker, Larimichthys crocea[J]. Journal of Fishery Sciences of China, 2018, 25(2): 361−372. doi: 10.3724/SP.J.1118.2018.17180 [17] 姜燕, 徐永江, 于超勇, 等. 大黄鱼消化道菌群结构、消化酶和非特异性免疫酶活力分析[J]. 渔业科学进展, 2020, 41(5): 61−72.Jiang Yan, Xu Yongjiang, Yu Chaoyong, et al. Analysis of microbiota structure, digestive enzyme and nonspecific immune enzyme activity in the gastrointestinal tract of large yellow croaker[J]. Progress in Fishery Sciences, 2020, 41(5): 61−72. [18] 姜燕, 于超勇, 徐永江, 等. 健康与患病大黄鱼消化道微生物结构特征分析[J]. 中国海洋大学学报(自然科学版), 2021, 51(5): 32−40.Jiang Yan, Yu Chaoyong, Xu Yongjiang, et al. Analysis of microbiota structural characteristic in the gastrointestinal tract of healthy and diseased large yellow croaker[J]. Periodical of Ocean University of China, 2021, 51(5): 32−40. [19] 崔龙波, 迟爽, 李新华, 等. 大黄鱼消化系统的组织学和组织化学研究[J]. 烟台大学学报(自然科学与工程版), 2014, 27(4): 266−270.Cui Longbo, Chi Shuang, Li Xinhua, et al. Histological and histochemistrical study on digestive system of Pseudosciaena crocea riciardson[J]. Journal of Yantai University (Natural Science and Engineering Edition), 2014, 27(4): 266−270. [20] Kokou F, Sasson G, Friedman J, et al. Core gut microbial communities are maintained by beneficial interactions and strain variability in fish[J]. Nature Microbiology, 2019, 4(12): 2456−2465. doi: 10.1038/s41564-019-0560-0 [21] Magoč T, Salzberg S L. FLASH: fast length adjustment of short reads to improve genome assemblies[J]. Bioinformatics, 2011, 27(21): 2957−2963. doi: 10.1093/bioinformatics/btr507 [22] Caporaso J G, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data[J]. Nature Methods, 2010, 7(5): 335−336. doi: 10.1038/nmeth.f.303 [23] Edgar R C, Haas B J, Clemente J C, et al. UCHIME improves sensitivity and speed of chimera detection[J]. Bioinformatics, 2011, 27(16): 2194−2200. doi: 10.1093/bioinformatics/btr381 [24] Edgar R C. UPARSE: highly accurate OTU sequences from microbial amplicon reads[J]. Nature Methods, 2013, 10(10): 996−998. doi: 10.1038/nmeth.2604 [25] Chen Xiaobing, Di Panpan, Wang Hongming, et al. Bacterial community associated with the intestinal tract of Chinese mitten crab ( Eriocheir sinensis) farmed in Lake Tai, China[J]. PLoS One, 2015, 10(4): e0123990. doi: 10.1371/journal.pone.0123990 [26] 孙健, 樊妙, 崔晓东, 等. 一种ReliefF和随机森林模型组合的多波束海底底质分类方法[J]. 海洋通报, 2022, 41(2): 131−139.Sun Jian, Fan Miao, Cui Xiaodong, et al. A multibeam seafloor classification method combining ReliefF and random forest model[J]. Marine Science Bulletin, 2022, 41(2): 131−139. [27] 万广, 陈忠辉, 方洪波, 等. 基于特征融合的随机森林模型茶鲜叶分类[J]. 华南农业大学学报, 2021, 42(4): 125−132.Wan Guang, Chen Zhonghui, Fang Hongbo, et al. Classification of fresh tea leaf based on random forest model by feature fusion[J]. Journal of South China Agricultural University, 2021, 42(4): 125−132. [28] 张昕雨, 张璟, 朱小强, 等. 基于宏基因组学分析构建诊断大肠癌的肠道菌群标签[J]. 上海交通大学学报(医学版), 2018, 38(9): 1019−1026.Zhang Xinyu, Zhang Jing, Zhu Xiaoqiang, et al. Bacterial signatures for diagnosis of colorectal cancer by fecal metagenomics analysis[J]. Journal of Shanghai Jiaotong University (Medical Science), 2018, 38(9): 1019−1026. [29] Xiong Jinbo, Nie Li, Chen Jiong. Current understanding on the roles of gut microbiota in fish disease and immunity[J]. Zoological Research, 2019, 40(2): 70−76. doi: 10.24272/j.issn.2095-8137.2018.069 [30] Zhu Jun, Li Hao, Jing Zezhou, et al. Robust host source tracking building on the divergent and non-stochastic assembly of gut microbiomes in wild and farmed large yellow croaker[J]. Microbiome, 2022, 10(1): 18. doi: 10.1186/s40168-021-01214-7 [31] 郁二蒙, 张振男, 夏耘, 等. 摄食不同饵料的大口黑鲈肠道菌群分析[J]. 水学学报, 2015, 39(1): 118−126.Yu Ermeng, Zhang Zhennan, Xia Yun, et al. Effects of different diets on intestinal microflora of largemouth bass ( Micropterus salmoides)[J]. Journal of Fisheries of China, 2015, 39(1): 118−126. [32] Xu Zhenjiang, Knight R. Dietary effects on human gut microbiome diversity[J]. British Journal of Nutrition, 2015, 113(S1): S1−S5. doi: 10.1017/S0007114514004127 [33] 陆振, 杨求华, 黄瑞芳, 等. 池塘养殖和海上吊笼养殖仿刺参肠道菌群结构对比分析[J]. 应用海洋学学报, 2017, 36(2): 187−194.Lu Zhen, Yang Qiuhua, Huang Ruifang, et al. Comparative analysis of intestinal microflora sea cucumber cultivated in ponds and sea suspension cages[J]. Journal of Applied Oceanography, 2017, 36(2): 187−194. [34] 盛鹏程, 周冬仁, 韩新荣, 等. 不同饲养方式对乌鳢( Ophiocephalus argus)肠道微生物群落结构差异及种类多样性的影响[J]. 海洋与湖沼, 2020, 51(1): 148−155.Sheng Pengcheng, Zhou Dongren, Han Xinrong, et al. Effects of different feeding matters on intestinal microbial structure diversity of Ophiocephalus argus[J]. Oceanologia et Limnologia Sinica, 2020, 51(1): 148−155. [35] Giatsis C, Sipkema D, Smidt H, et al. The impact of rearing environment on the development of gut microbiota in tilapia larvae[J]. Scientific Reports, 2015, 5(1): 18206. doi: 10.1038/srep18206 [36] Hennersdorf P, Mrotzek G, Abdul-Aziz M A, et al. Metagenomic analysis between free-living and cultured Epinephelus fuscoguttatus under different environmental conditions in Indonesian waters[J]. Marine Pollution Bulletin, 2016, 110(2): 726−734. doi: 10.1016/j.marpolbul.2016.05.009 [37] 苟妮娜, 钟明智, 王开锋. 基于16S rRNA高通量测序的野生和养殖多鳞白甲鱼肠道微生物群落组成研究[J]. 西北农业学报, 2021, 30(7): 963−970.Gou Nina, Zhong Mingzhi, Wang Kaifeng. Intestinal microbial community of wild and cultured Onychostoma macrolepi based on 16S rRNA high-throughput sequencing[J]. Acta Agriculturae Boreali-occidentalis Sinica, 2021, 30(7): 963−970. [38] Wu Jianfeng, Wen X W, Faulk C, et al. Perinatal lead exposure alters gut microbiota composition and results in sex-specific bodyweight increases in adult mice[J]. Toxicological Sciences, 2016, 151(2): 324−333. doi: 10.1093/toxsci/kfw046 [39] Aron-Wisnewsky J, Gaborit B, Dutour A, et al. Gut microbiota and non-alcoholic fatty liver disease: new insights[J]. Clinical Microbiology and Infection, 2013, 19(4): 338−348. doi: 10.1111/1469-0691.12140 [40] Schwiertz A, Taras D, Schäfer K, et al. Microbiota and SCFA in lean and overweight healthy subjects[J]. Obesity, 2010, 18(1): 190−195. doi: 10.1038/oby.2009.167 [41] Zhang Yuan, Wen Bin, David M A, et al. Comparative analysis of intestinal microbiota of discus fish ( Symphysodon haraldi) with different growth rates[J]. Aquaculture, 2021, 540: 736740. doi: 10.1016/j.aquaculture.2021.736740 [42] Gibiino G, Lopetuso L R, Scaldaferri F, et al. Exploring Bacteroidetes: metabolic key points and immunological tricks of our gut commensals[J]. Digestive and Liver Disease, 2018, 50(7): 635−639. doi: 10.1016/j.dld.2018.03.016 [43] Shin N R, Whon T W, Bae J W. Proteobacteria: microbial signature of dysbiosis in gut microbiota[J]. Trends in Biotechnology, 2015, 33(9): 496−503. doi: 10.1016/j.tibtech.2015.06.011 [44] 李英英, 陈曦, 宋铁英. 不同生长速度的大黄鱼肠道菌群结构的差异[J]. 大连海洋大学学报, 2017, 32(5): 509−513. doi: 10.16535/j.cnki.dlhyxb.2017.05.002Li Yingying, Chen Xi, Song Tieying. Differences in intestinal flora of cultured large yellow croaker Pseudosciaena crocea with different growth rates[J]. Journal of Dalian Ocean University, 2017, 32(5): 509−513. doi: 10.16535/j.cnki.dlhyxb.2017.05.002 [45] 李欣海. 随机森林模型在分类与回归分析中的应用[J]. 应用昆虫学报, 2013, 50(4): 1190−1197.Li Xinhai. Using “random forest” for classification and regression[J]. Chinese Journal of Applied Entomology, 2013, 50(4): 1190−1197. -




 下载:
下载: