Effects of CO2-driven seawater acidification on tissue, immune and antioxidant enzyme activity and transcription levels of Ruditapes philippinarum
-
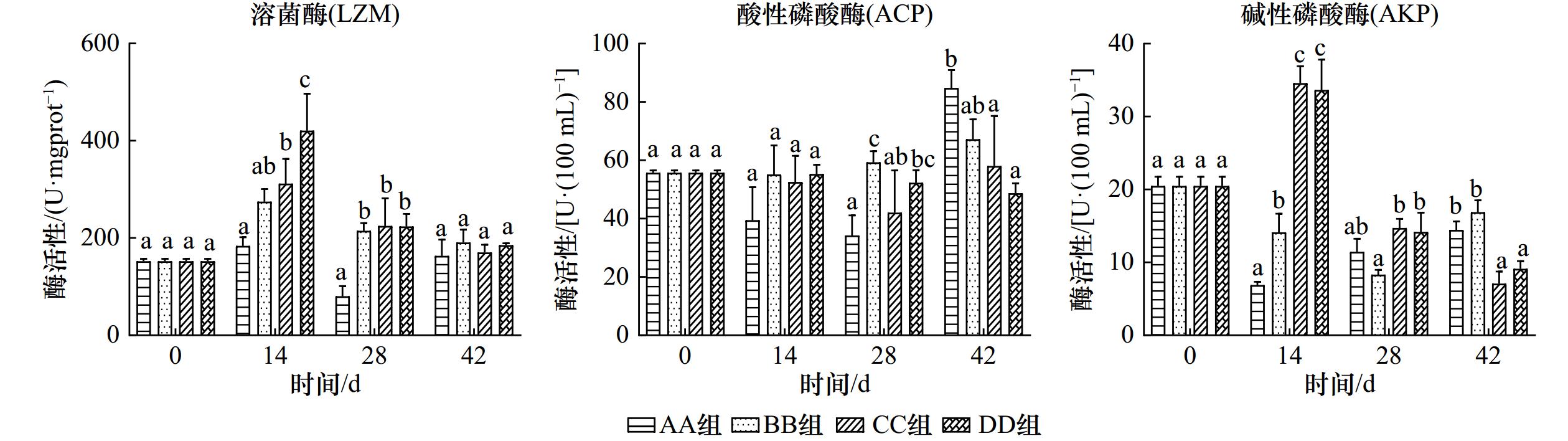
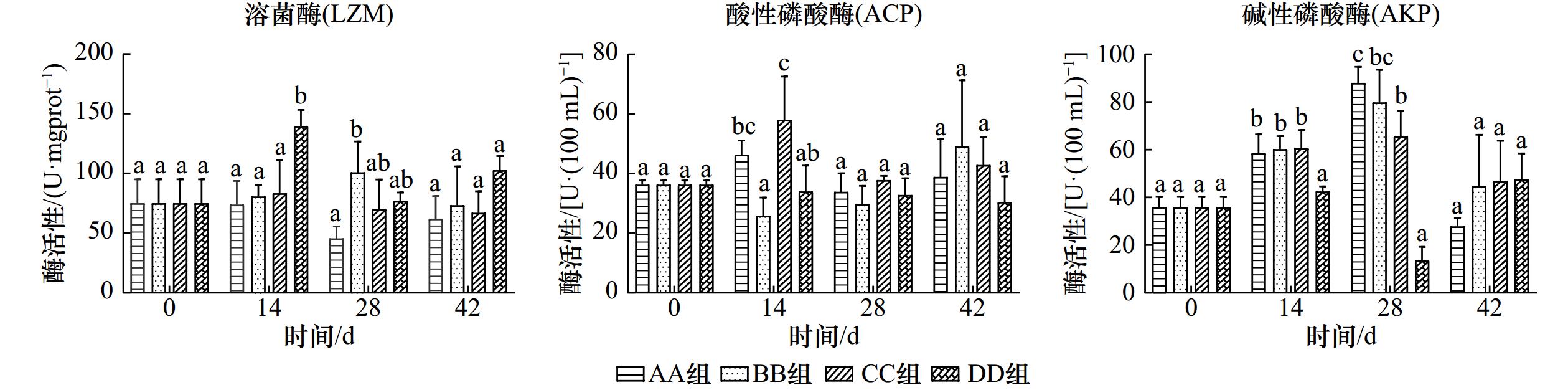
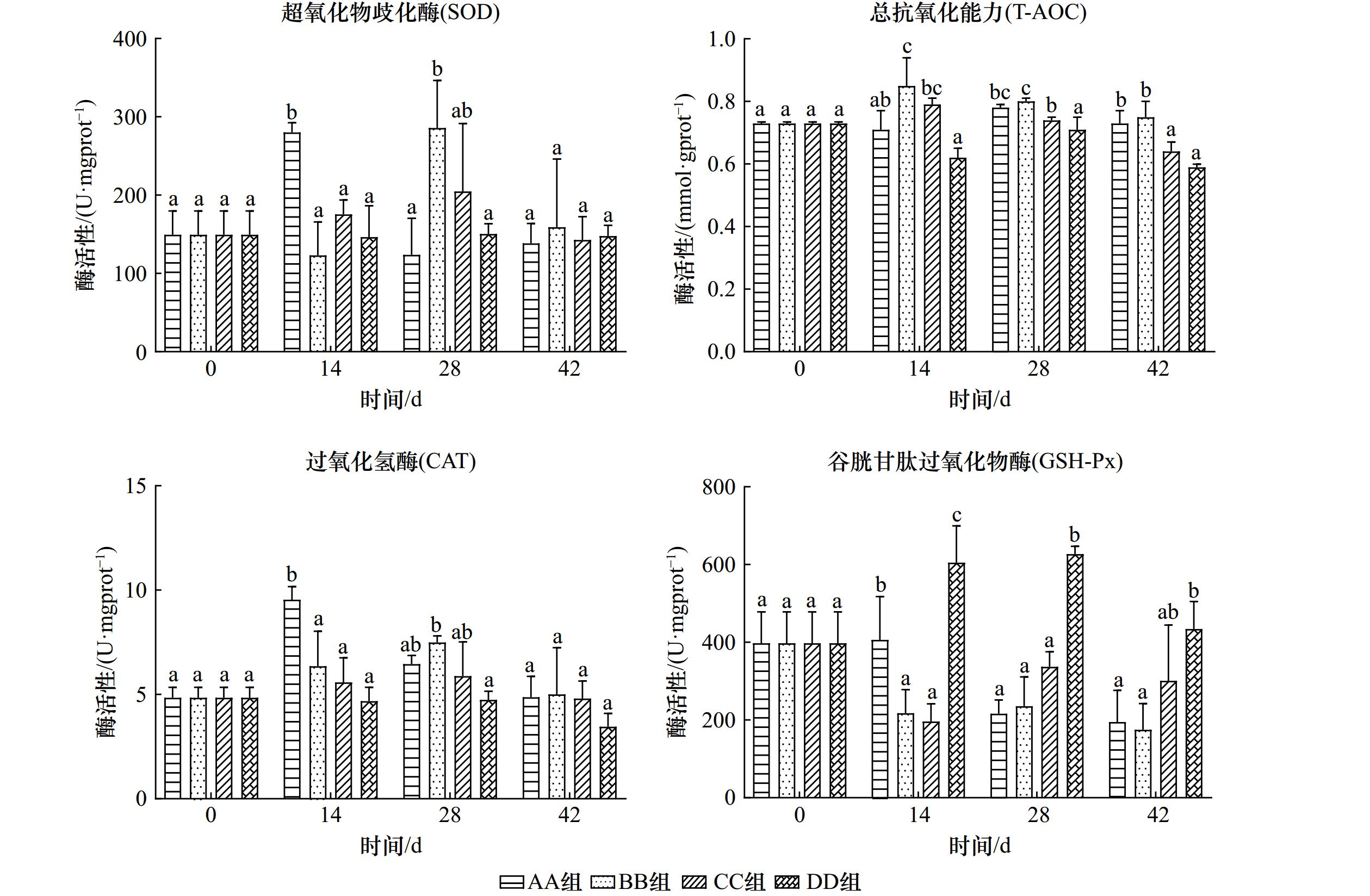
摘要: 随着CO2的大量排放,海洋酸化效应不断加重,为探究未来海水酸化情况对菲律宾蛤仔产生的影响,设置对照组(pH为8.1)和酸化组(pH为7.7、7.1和6.4),研究周期为42 d,测定菲律宾蛤仔在酸化条件下组织结构、免疫和抗氧化酶活性的变化情况,以及在分子水平上产生的影响。结果表明:菲律宾蛤仔置于酸化海水环境中,鳃丝间距随pH的降低而扩大,鳃丝纤毛黏合,水管和外套膜外表皮褶皱逐渐加深;鳃组织中酸性磷酸酶(ACP)和超氧化物歧化酶(SOD)活性变化情况为先降后升,碱性磷酸酶(AKP)活性各组变化趋势不同,总抗氧化能力(T-AOC)、过氧化氢酶(CAT)和溶菌酶(LZM)活性趋势为先升后降;鳃和内脏团谷胱甘肽过氧化物酶(GSH-Px)活性变化规律皆为持续上升;内脏团组织中LZM活性变化趋势各不相同,ACP活性变动趋势为先降后升,AKP、SOD和CAT活性变化规律为先升后降,T-AOC趋势为持续下降;通过转录组的分析得到,鳃组织GO功能主要富集在DNA整合、膜的组成部分和RNA定向DNA聚合酶活性等条目中,KEGG通路主要富集在吞噬体和与蛋白合成的相关通路中。海水酸化使菲律宾蛤仔组织呈现不同程度的损伤,破坏其内环境稳态,改变其代谢水平和与免疫相关基因表达,引发菲律宾蛤仔染病乃至死亡的风险上升。Abstract: The ocean acidification effect is increasing with the large amount of CO2 emissions. To investigate the effects of future seawater acidification on Ruditapes philippinarum, a control group (pH = 8.1) and acidification group (pH = 7.7, 7.1 and 6.4) were set up for 42 days. The changes in tissue structure, immune and antioxidant enzyme activities of Ruditapes philippinarum under acidification conditions were measured, as well as the effects produced at the molecular level. The results show that when Ruditapes philippinarum are placed in an acidified seawater environment, gill filament spacing expands with decreasing pH, gill filament cilia adhere, and the pipes and outer epidermal folds of the mantle gradually deepen. The activities of acid phosphatase (ACP) and superoxide dismutase (SOD) in gill tissues show a pattern of decreasing followed by increasing. Alkaline phosphatase (AKP) activities exhibit different trends in each group. Total antioxidant capacity (T-AOC), catalase (CAT), and lysozyme (LZM) activities show a pattern of increasing followed by decreasing. Glutathione peroxidase (GSH-Px) activities in gill and visceral masses show a continuous increase. LZM activity in the viscera group displays varying trends, while ACP activity shows a decreasing and then increasing pattern. AKP, SOD, and CAT activities exhibited an increasing and then decreasing pattern, while T-AOC activity shows a continuous decrease. Analysis of the transcriptome reveals that the GO functions in gill tissue are mainly enriched in DNA integration, integral components of the membrane, and RNA-directed DNA polymerase activity, among others. The KEGG pathway analysis shows enrichment in the phagosome pathway and pathways related to protein processing in the endoplasmic reticulum. The acidification of seawater caused varying degrees of damage to the tissues of Ruditapes philippinarum, disrupting its internal environmental homeostasis and altering metabolic levels and immune-related gene expression, and led to an increased risk of disease and even death in Ruditapes philippinarum.
-
Key words:
- carbon dioxide /
- Ruditapes philippinarum /
- immunity /
- antioxidant /
- transcript levels
-
图 1 海水酸化对菲律宾蛤仔不同组织结构的影响
A. DD组鳃;B. CC组鳃;C. BB组鳃;D. AA组鳃;E. DD组水管;F. CC组水管;G. BB组水管;H. AA组水管;I. DD组外套膜;J. CC组外套膜;K. BB组外套膜;L. AA组外套膜
Fig. 1 Effects of seawater acidification on different organization structure of Ruditapes philippinarum
A. Group DD gill; B. Group CC gill; C. Group BB gill; D. Group AA gill; E. Group DD pipe; F. Group CC pipe; G. Group BB pipe; H. Group AA pipe; I. Group DD mantle; J. Group CC mantle; K. Group BB mantle; L. Group AA mantle
表 1 菲律宾蛤仔鳃组织文库测序数据统计分析
Tab. 1 Statistical analysis of sequencing data of gill library of Ruditapes philippinarum
样品 序列数 质控数据总碱基数/bp GC含量/% Q20/% Q30/% AA1 19 805 256 5 921 771 252 36.81 97.42 92.53 AA2 21 015 864 6 280 860 796 36.94 97.54 92.78 AA3 19 731 774 5 901 090 298 37.18 97.5 92.68 BB1 20 674 908 6 180 447 036 37.45 97.76 93.27 BB2 20 510 382 6 131 925 988 37.4 96.53 90.69 BB3 20 992 198 6 275 544 498 35.81 97.11 91.92 CC1 21 890 314 6 542 576 532 36.44 97.11 91.92 CC2 26 002 371 7 775 975 502 36.79 97.55 92.78 CC3 19 225 650 5 747 607 418 36.92 97.76 93.31 DD1 26 236 765 7 799 003 780 37.5 97.66 93.08 DD2 21 474 785 6 390 329 892 36.4 97.52 92.79 DD3 20 597 574 6 154 866 696 36.74 97.27 92.24 表 2 差异表达基因GO功能富集
Tab. 2 GO functional enrichment of differentially expressed genes
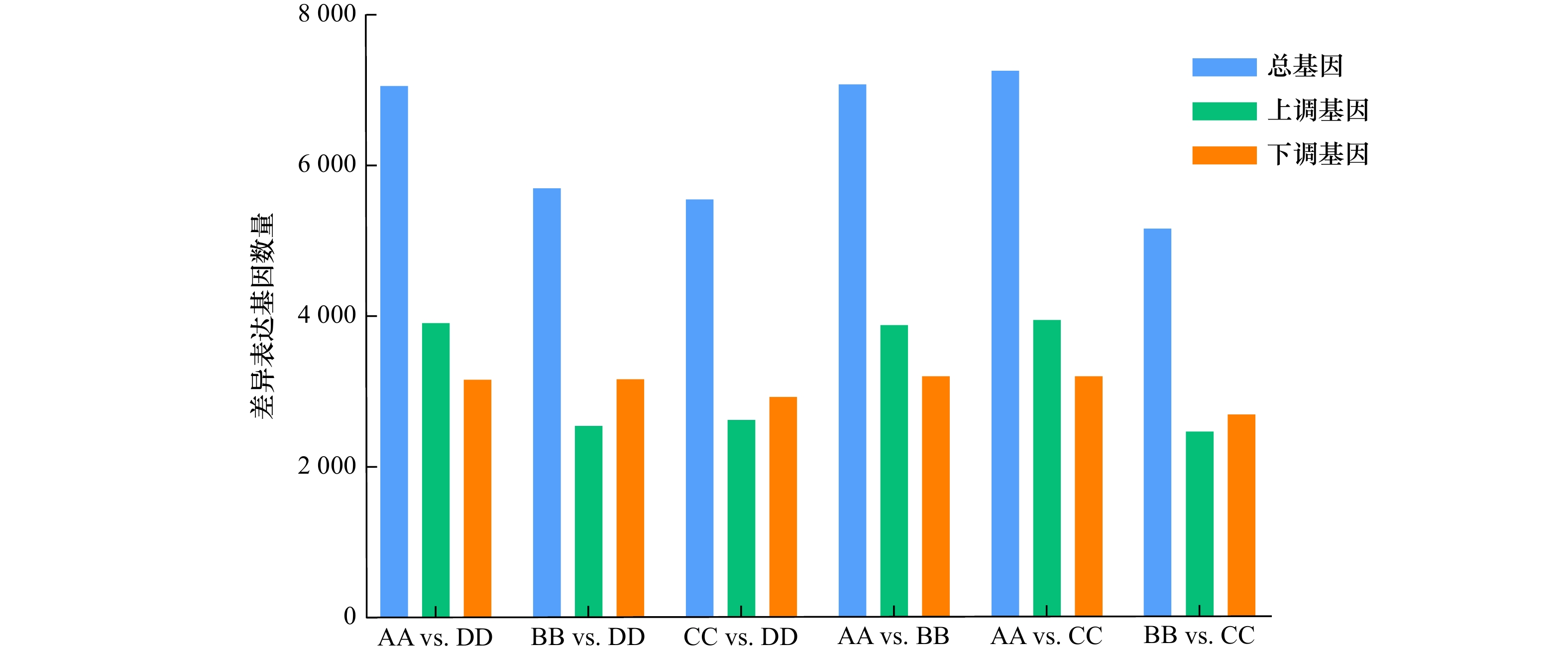
差异表达基因类型 GO条目 p值 AA vs. DD BB vs. DD CC vs. DD AA vs. BB AA vs. CC BB vs. CC 上调基因 DNA整合 1.48 × 10−5 1.61 × 10−10 1.84 × 10−4 3.79 × 10−5 5.59 × 10−5 5.03 × 10−9 膜的组成部分 1.85 × 10−2 5.85 × 10−4 0.43 1.13 × 10−2 5.07 × 10−4 1.04 × 10−2 核糖体 5.99 × 10−5 0.92 1.37 × 10−2 1.49 × 10−2 1.54 × 10−4 0.14 RNA定向DNA聚合酶活性 0.59 1.24 × 10−2 1.17 × 10−4 1.93 × 10−5 7.16 × 10−2 1.52 × 10−4 下调基因 包涵体组装的负调节 4.74 × 10−5 0.70 0.34 4.79 × 10−5 3.45 × 10−4 0.68 蛋白水解 5.32 × 10−5 3.70 × 10−7 6.20 × 10−6 2.48 × 10−2 5.14 × 10−2 9.56 × 10−2 细胞外区域 1.61 × 10−11 8.53 × 10−12 1.83 × 10−4 2.79 × 10−12 4.18 × 10−14 1.55 × 10−9 细胞外空间 1.56 × 10−8 1.57 × 10−5 2.52 × 10−2 5.41 × 10−7 1.36 × 10−5 2.79 × 10−4 内肽酶抑制剂活性 2.06 × 10−7 7.27 × 10−8 1.16 × 10−7 6.57 × 10−2 0.29 2.26 × 10−3 表 3 差异表达基因KEGG信号通路富集
Tab. 3 KEGG pathways enriched for differentially expressed genes
差异表达基因类型 通路名称 基因数目 AA vs. DD BB vs. DD CC vs. DD AA vs. BB AA vs. CC BB vs. CC 上调基因 真核生物中的核糖体生物发生 41 6 1 33 45 6 核糖体 85 25 42 57 76 33 Notch信号传导途径通路 18 21 15 17 15 15 NOD-like受体信号通路 13 16 13 17 19 15 泛素介导的蛋白水解 32 21 20 44 43 26 下调基因 吞噬体 41 25 20 42 35 20 内质网中的蛋白加工 12 7 8 21 14 2 细胞外基质受体作用 17 16 10 23 16 8 C-型凝集素受体信号通路 14 7 9 13 19 14 -
[1] Sabine C L, Feely R A, Gruber N, et al. The oceanic sink for anthropogenic CO2[J]. Science, 2004, 305(5682): 367−371. doi: 10.1126/science.1097403 [2] 石莉, 桂静, 吴克勤. 海洋酸化及国际研究动态[J]. 海洋科学进展, 2011, 29(1): 122−128.Shi Li, Gui Jing, Wu Keqin. Developments in international studies on ocean acidification[J]. Advances in Marine Science, 2011, 29(1): 122−128. [3] Orr J C, Fabry V J, Aumont O, et al. Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms[J]. Nature, 2005, 437(7059): 681−686. doi: 10.1038/nature04095 [4] Caldeira K, Wickett M E. Ocean model predictions of chemistry changes from carbon dioxide emissions to the atmosphere and ocean[J]. Journal of Geophysical Research: Oceans, 2005, 110(C9): C09S04. [5] 常亚青. 贝类增养殖学[M]. 北京: 中国农业出版社, 2007.Chang Yaqing. Science of Shellfish Cultivation[M]. Beijing: China Agriculture Press, 2007. [6] 农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会. 2022中国渔业统计年鉴[M]. 北京: 中国农业出版社, 2022.Fisheries and Fisheries Administration Bureau of the Ministry of Agriculture and Rural Affairs, National Fisheries Technology Extension Center, China Society of Fisheries. 2022 China Fishery Statistical Yearbook[M]. Beijing: China Agriculture Press, 2022. [7] Gazeau F, Quiblier C, Jansen J M, et al. Impact of elevated CO2 on shellfish calcification[J]. Geophysical Research Letters, 2007, 34(7): L07603. [8] Talmage S C, Gobler C J. Effects of past, present, and future ocean carbon dioxide concentrations on the growth and survival of larval shellfish[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(40): 17246−17251. [9] Bibby R, Widdicombe S, Parry H, et al. Effects of ocean acidification on the immune response of the blue mussel Mytilus edulis[J]. Aquatic Biology, 2008, 2(1): 67−74. [10] Love M I, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biology, 2014, 15(12): 550. doi: 10.1186/s13059-014-0550-8 [11] Yu Guangchuang, Wang Ligen, Han Yanyan, et al. clusterProfiler: an R package for comparing biological themes among gene clusters[J]. OMICS: A Journal of Integrative Biology, 2012, 16(5): 284−287. doi: 10.1089/omi.2011.0118 [12] 徐宏超, 邢荣莲, 李源美, 等. 氨氮胁迫下菲律宾蛤仔肝胰腺内参基因的筛选[J]. 生态毒理学报, 2021, 16(3): 218−226.Xu Hongchao, Xing Ronglian, Li Yuanmei, et al. Screening of reference genes in hepatopancreas of clam Ruditapes philippinarum exposed to ammonia nitrogen[J]. Asian Journal of Ecotoxicology, 2021, 16(3): 218−226. [13] 宋晓楠, 马峻峰, 秦艳杰, 等. 盐度骤降对菲律宾蛤仔抗氧化酶活力及组织结构的影响[J]. 农学学报, 2013, 3(1): 50−56, 70.Song Xiaonan, Ma Junfeng, Qin Yanjie, et al. Effects of abrupt decline in salinity on the antioxidant enzyme activities and histological structure in Ruditapes philippenarum[J]. Journal of Agriculture, 2013, 3(1): 50−56, 70. [14] 梁健, 朱飞霞, 刘宇航, 等. 海洋酸化对缢蛏鳃组织结构的影响[J]. 经济动物学报, 2022, 26(4): 299−303.Liang Jian, Zhu Feixia, Liu Yuhang, et al. Effects of ocean acidification on gill structure of razor clams Sinonovacula constricta[J]. Journal of Economic Animal, 2022, 26(4): 299−303. [15] 周瑶, 刘丽丹, 孙庆业. 煤矸石对褶纹冠蚌斧足和外套膜重金属含量及组织结构的影响[J]. 西南农业学报, 2022, 35(1): 250−256.Zhou Yao, Liu Lidan, Sun Qingye. Effects of coal gangue on heavy metal content and tissue structure of foot and mantellum of Cristaria plicata[J]. Southwest China Journal of Agricultural Sciences, 2022, 35(1): 250−256. [16] 孙虎山, 李光友. 脂多糖对栉孔扇贝血清和血细胞中7种酶活力的影响[J]. 海洋科学, 1999, 23(4): 54−58.Sun Hushan, Li Guangyou. Effects of lipopolysaccharide on enzymes in serum and haemocytes of Chlamys farreri[J]. Marine Sciences, 1999, 23(4): 54−58. [17] 郑萍萍, 王春琳, 宋微微, 等. 盐度胁迫对三疣梭子蟹血清非特异性免疫因子的影响[J]. 水产科学, 2010, 29(11): 634−638.Zheng Pingping, Wang Chunlin, Song Weiwei, et al. Effect of salinity stress on serum non-specific immune factors in swimming crab Portunus trituberculatus[J]. Fisheries Science, 2010, 29(11): 634−638. [18] 房子恒, 田相利, 董双林, 等. 不同盐度下半滑舌鳎幼鱼非特异性免疫酶活力分析[J]. 中国海洋大学学报(自然科学版), 2014, 44(5): 46−53.Fang Ziheng, Tian Xiangli, Dong Shuanglin, et al. Analysis of the activity of non-specific immune enzymes of juvenile tongue soles cultured in various salinities[J]. Periodical of Ocean University of China, 2014, 44(5): 46−53. [19] 谢莉萍, 林静瑜, 肖锐, 等. 合浦珠母贝碱性磷酸酶的分离纯化与性质研究[J]. 海洋科学, 2000, 24(10): 37−40. doi: 10.3969/j.issn.1000-3096.2000.10.014Xie Liping, Lin Jingyu, Xiao Rui, et al. Purification and characterization of alkaline phosphatase from Pinctada fucata[J]. Marine Sciences, 2000, 24(10): 37−40. doi: 10.3969/j.issn.1000-3096.2000.10.014 [20] 刘璐, 杨玉娇, 王国良. pH突变对拟穴青蟹免疫因子的胁迫影响[J]. 宁波大学学报(理工版), 2009, 22(4): 484−489.Liu Lu, Yang Yujiao, Wang Guoliang. Effect of sudden changes in pH on the immune factors of Scylla paramamosain[J]. Journal of Ningbo University (Natural Science & Engineering Edition), 2009, 22(4): 484−489. [21] 刘树青, 江晓路, 牟海津, 等. 免疫多糖对中国对虾血清溶菌酶、磷酸酶和过氧化物酶的作用[J]. 海洋与湖沼, 1999, 30(3): 278−283.Liu Shuqing, Jiang Xiaolu, Mou Haijin, et al. Effects of immunopoiysaccharide on LSZ, ALP, ACP and POD activities of Penaeus Chinensis serum[J]. Oceanologia et Limnologia Sinica, 1999, 30(3): 278−283. [22] 赵丹, 周丽青, 吴彪, 等. 魁蚶各组织溶菌酶活性对鳗弧菌侵染的响应[J]. 水产学报, 2020, 44(3): 480−486.Zhao Dan, Zhou Liqing, Wu Biao, et al. Response of lysozyme activity to Vibrio anguillarum infection in different tissues of Scapharca broughtonii[J]. Journal of Fisheries of China, 2020, 44(3): 480−486. [23] 姜令绪, 潘鲁青, 肖国强. 氨氮对凡纳对虾免疫指标的影响[J]. 中国水产科学, 2004, 11(6): 537−541. doi: 10.3321/j.issn:1005-8737.2004.06.009Jiang Lingxu, Pan Luqing, Xiao Guoqiang. Effects of ammonia-N on immune parameters of white shrimp Litopenaeus vannamei[J]. Journal of Fishery Sciences of China, 2004, 11(6): 537−541. doi: 10.3321/j.issn:1005-8737.2004.06.009 [24] 孙敬锋, 吴信忠. 贝类血细胞及其免疫功能研究进展[J]. 水生生物学报, 2006, 30(5): 601−607.Sun Jingfeng, Wu Xinzhong. The progress of studies on molluscan hemocyte and its immunological function[J]. Acta Hydrobiologica Sinica, 2006, 30(5): 601−607. [25] 樊甄姣, 杨爱国, 刘志鸿, 等. pH对栉孔扇贝体内几种免疫因子的影响[J]. 中国水产科学, 2006, 13(4): 650−654.Fan Zhenjiao, Yang Aiguo, Liu Zhihong, et al. Effect of pH on the immune factors of Chlamys farreri[J]. Journal of Fishery Sciences of China, 2006, 13(4): 650−654. [26] 潘红春, 范杰, 王芳芳, 等. pH值对日本三角涡虫种群增长、无性生殖及6种酶活力的影响[J]. 水生生物学报, 2008, 32(3): 339−344. doi: 10.3724/SP.J.1035.2008.00339Pan Hongchun, Fan Jie, Wang Fangfang, et al. Effect of medium pH on population growth, asexual reproduction and activity of six enzymes of Dugesia japonica[J]. Acta Hydrobiologica Sinica, 2008, 32(3): 339−344. doi: 10.3724/SP.J.1035.2008.00339 [27] 李晨辉, 王梦杰, 彭艳青, 等. 海洋酸化对脊尾白虾抗氧化酶活性的影响[J]. 淮海工学院学报(自然科学版), 2018, 27(4): 78−83.Li Chenhui, Wang Mengjie, Peng Yanqing, et al. Effect of ocean acidification on activities of antioxidant enzymein Exopalaemon carinicauda[J]. Journal of Jiangsu Ocean University (Natural Science Edition), 2018, 27(4): 78−83. [28] Gaetani G F, Ferraris A M, Rolfo M, et al. Predominant role of catalase in the disposal of hydrogen peroxide within human erythrocytes[J]. Blood, 1996, 87(4): 1595−1599. doi: 10.1182/blood.V87.4.1595.bloodjournal8741595 [29] Tiedke J, Cubuk C, Burmester T. Environmental acidification triggers oxidative stress and enhances globin expression in zebrafish gills[J]. Biochemical and Biophysical Research Communications, 2013, 441(3): 624−629. doi: 10.1016/j.bbrc.2013.10.104 [30] Apel K, Hirt H. REACTIVE OXYGEN SPECIES: metabolism, oxidative stress, and signal transduction[J]. Annual Review of Plant Biology, 2004, 55: 373−399. doi: 10.1146/annurev.arplant.55.031903.141701 [31] Warner H R. Superoxide dismutase, aging, and degenerative disease[J]. Free Radical Biology and Medicine, 1994, 17(3): 249−258. doi: 10.1016/0891-5849(94)90080-9 [32] Villar-Piqué A, Ventura S. Protein aggregation propensity is a crucial determinant of intracellular inclusion formation and quality control degradation[J]. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 2013, 1833(12): 2714−2724. doi: 10.1016/j.bbamcr.2013.06.023 [33] Theocharis A D, Skandalis S S, Gialeli C, et al. Extracellular matrix structure[J]. Advanced Drug Delivery Reviews, 2016, 97: 4−27. doi: 10.1016/j.addr.2015.11.001 [34] Wang Le, Sun Fei, Wen Yanfei, et al. Effects of ocean acidification on transcriptomes in Asian seabass juveniles[J]. Marine Biotechnology, 2021, 23(3): 445−455. doi: 10.1007/s10126-021-10036-5 [35] 周岳华, 黄勇, 翟玲, 等. 真核生物核糖体和mRNA翻译的研究进展[J]. 安徽农业科学, 2012, 40(24): 11946−11948. doi: 10.3969/j.issn.0517-6611.2012.24.007Zhou Yuehua, Huang Yong, Zhai Ling, et al. Research progress in eukaryotic ribosome and mRNA translation[J]. Journal of Anhui Agricultural Sciences, 2012, 40(24): 11946−11948. doi: 10.3969/j.issn.0517-6611.2012.24.007 [36] 贾若南, 林枫, 许强华. 低氧胁迫下斑马鱼鳃中核糖体蛋白基因家族的表达分析[J]. 上海海洋大学学报, 2022, 31(2): 318−327.Jia Ruonan, Lin Feng, Xu Qianghua. Differential expression analysis of the ribosomal protein gene family in zebrafish gills under hypoxia stress[J]. Journal of Shanghai Ocean University, 2022, 31(2): 318−327. [37] Henrique D, Schweisguth F. Mechanisms of Notch signaling: a simple logic deployed in time and space[J]. Development, 2019, 146(3): dev172148. doi: 10.1242/dev.172148 [38] Zheng Chunfu. The emerging roles of NOD-like receptors in antiviral innate immune signaling pathways[J]. International Journal of Biological Macromolecules, 2021, 169: 407−413. doi: 10.1016/j.ijbiomac.2020.12.127 [39] Johnson K M, Hofmann G E. A transcriptome resource for the Antarctic pteropod Limacina helicina antarctica[J]. Marine Genomics, 2016, 28: 25−28. doi: 10.1016/j.margen.2016.04.002 [40] 刘俊文, 杨向东. 泛素−蛋白酶体途径介导的细胞周期调节蛋白的水解[J]. 国外医学·老年医学分册, 2005, 26(1): 30−34.Liu Junwen, Yang Xiangdong. Ubiquitin-proteasome pathway mediated hydrolysis of cell cycle regulatory proteins[J]. International Journal of Geriatrics, 2005, 26(1): 30−34. [41] Zhang Dongsheng, Yu Mengchao, Hu Peng, et al. Genetic adaptation of Schizothoracine fish to the phased uplifting of the Qinghai-Tibetan Plateau[J]. G3-Genes Genomes Genetics, 2017, 7(4): 1267−1276. [42] Todgham A E, Crombie T A, Hofmann G E. The effect of temperature adaptation on the ubiquitin-proteasome pathway in notothenioid fishes[J]. Journal of Experimental Biology, 2017, 220(3): 369−378. [43] Sutherland T E, Dyer D P, Allen J E. The extracellular matrix and the immune system: a mutually dependent relationship[J]. Science, 2023, 379(6633): eabp8964. doi: 10.1126/science.abp8964 [44] Hatinguais R, Willment J A, Brown G D. C-type lectin receptors in antifungal immunity: current knowledge and future developments[J]. Parasite Immunology, 2023, 45(2): e12951. doi: 10.1111/pim.12951 -





 下载:
下载: