Composition and distribution of methane metabolic archaea in oceanic surface sediments
-
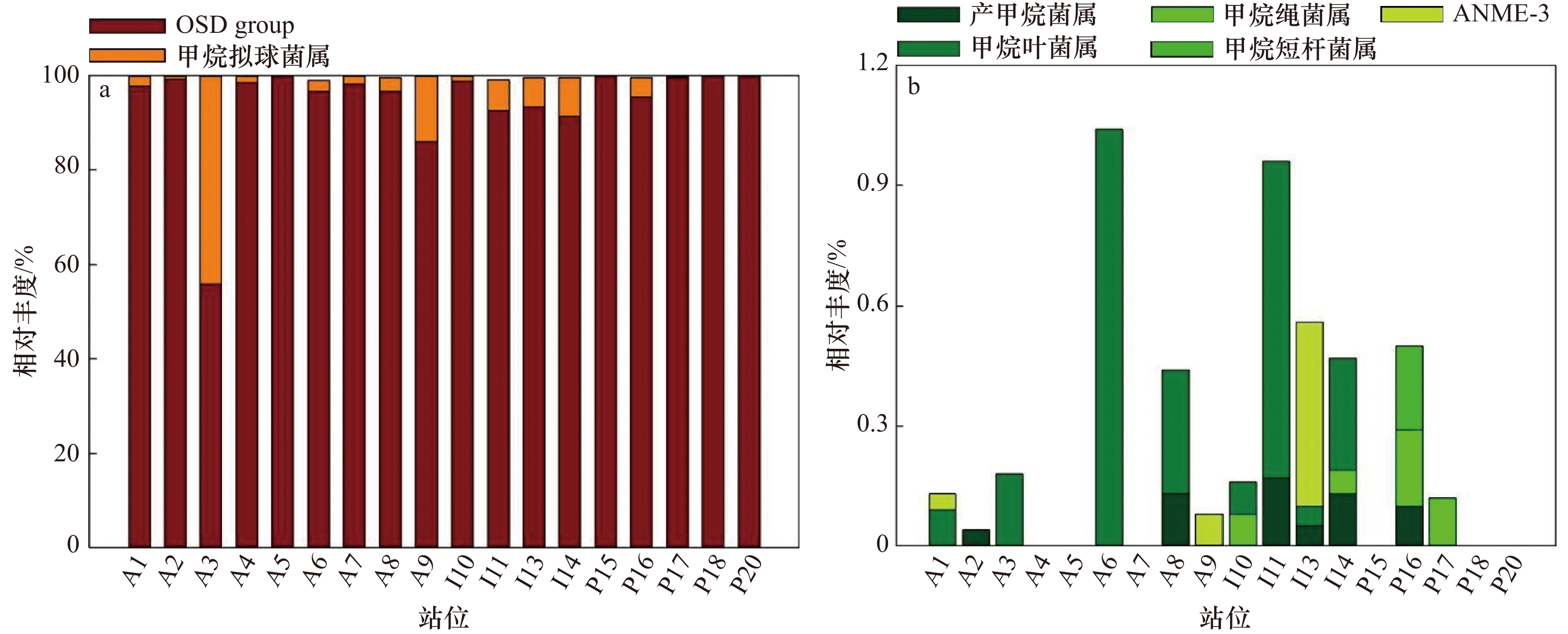
摘要: 海洋沉积物中的甲烷代谢微生物是甲烷循环的关键参与者,其代谢过程对大气甲烷浓度及全球气候变化具有显著影响,研究其在全球大洋沉积物中的组成及分布特征是探究微生物介导甲烷循环的基础。采用焦磷酸454高通量测序测定甲烷代谢保守功能基因mcrA(Methyl coenzyme–M reductase A)分析全球大洋沉积物中甲烷代谢微生物群落的组成和多样性;结合荧光实时定量PCR技术检测了古菌和甲烷代谢古菌的丰度分布特征。与其他海洋生境对比,大洋沉积物中甲烷代谢古菌群落结构单一,大西洋和印度洋的α多样性指数显著高于太平洋(p<0.05)。在大洋沉积物样品中鉴定到3个目的甲烷代谢古菌,即甲烷杆菌目(Methanobacteriales)、甲烷八叠球菌目(Methanosarcinales)和甲烷微菌目(Methanomicrobiales),其中甲烷微菌目占绝对优势,并主要由一簇未知类群(暂名Oceanic Sediments Dominant group,OSD group)组成。大洋沉积物的古菌16S rRNA基因丰度(湿重,下同)平均为8.81×106 copies/g,大西洋的低于印度洋和太平洋;马里亚纳海沟基因丰度低于南海北部,且随着采样深度增加而呈降低趋势。大洋沉积物的mcrA基因丰度为1.38×103~8.25×104 copies/g。基因丰度大西洋最高,太平洋次之,印度洋最低;马里亚纳海沟略高于南海。本研究发现,相较于冷泉、热液、近海河口等海洋生境,大洋沉积物中甲烷代谢古菌丰度低且群落结构单一,不同海区样品间具有极高的相似性;同时发现OSD group是全球大洋沉积物样品的绝对优势类群,其与已知类群序列亲缘关系均较远,分类进化地位尚不明晰,值得进一步研究。Abstract: The microorganisms that generate and utilize methane are recognized as the key parts of global methane cycle, and their metabolic process has significant effects on global climate change. The knowledge upon composition and distribution characteristics of these methane metabolic microorganisms in global ocean sediments is the basis to understand the participation of methane metabolizing microorganisms in marine sediments in methane cycle. Pyrosequencing based on functional gene mcrA (methyl coenzyme-M reductase A) was applied to study the community composition and abundance of methane metabolic microbes in global ocean sediments, and their abundance together with those of archaea were estimated with fluorescence real-time quantitative PCR. Compared with other marine habitats, the methane metabolic archaeal communities in the deep ocean sediments had a much lower diversity and simpler community structure. The α-diversity in Atlantic Ocean and Indian Ocean was significantly higher than that in the Pacific Ocean (p<0.05). Methanogens revealed from this study belong to three orders, Methanobacteriales, Methanosarcinales, and Methanomicrobiales. Methanomicrobiales as the dominant one was predominated by an unknown OSD group. The average abundance (wet weight, the same below) of archaeal 16S rRNA gene in all sediments was 8.81×106 copies/g. Those in Atlantic Ocean was lower than the other two oceans; while those in the Mariana Trench was lower than in the northern part of the South China Sea, and tended to decrease with the increase of sediment depth. The abundance of mcrA gene in all sediment samples was 1.38×103–8.25×104 copies/g. Those in Atlantic Ocean was the highest. It revealed that low abundance and diversity of methane metabolic archaea in the deep-sea sediments, and the high similarities among different communities among samples in different oceans. In addition, the OSD group recovered as the predominant group in this study had no close relationship with any other known genotypes in the GenBank, therefore, its taxonomic status was unknown and deserve further study.
-
表 1 沉积物样品的采样环境参数
Tab. 1 Sampling parameters in different ocean sediments
编号 海区 纬度 经度 站位深度/m 采样时间 A1 南大西洋中脊 13.589 6°S 14.519 3°W 3 203 2012年7月25日 A2 南大西洋中脊 13.355 1°S 14.311 1°W 3 142 2012年7月26日 A3 南大西洋中脊 13.593 5°S 14.519 2°W 3 149 2012年7月27日 A4 南大西洋中脊 13.594 6°S 14.517 7°W 3 073 2012年7月29日 A5 南大西洋中脊 14.056 6°S 14.354 8°W 1 598 2012年8月8日 A6 南大西洋中脊 19.347 3°S 11.923 2°W 2 597 2012年9月25日 A7 南大西洋中脊 19.260 7°S 11.919 8°W 2 621 2012年9月27日 A8 南大西洋中脊 19.408 5°S 11.885 1°W 2 400 2012年9月27日 A9 南大西洋中脊 19.260 7°S 11.930 2°W 2 559 2012年9月30日 I10 西北印度洋脊 3.710 6°N 63.657 3°E 3 690 2013年4月24日 I11 西北印度洋脊 5.716 8°N 61.462 1°E 4 071 2013年4月28日 I12 西北印度洋脊 6.363 2°N 60.525 5°E 2 989 2013年5月1日 I13 西北印度洋脊 9.066 1°N 58.252 7°E 2 844 2013年5月6日 I14 西北印度洋脊 9.774 7°N 57.581 9°E 2 088 2013年5月9日 P15 西太平洋 11.006 5°N 141.952 2°E 7 015 2012年6月30日 P16 西太平洋 11.006 5°N 141.952 2°E 7 015 2012年6月30日 P17 西太平洋 11.006 5°N 141.952 2°E 7 015 2012年6月30日 P18 西太平洋 18.989 1°N 113.924 1°E 1 015 2015年6月29日 P19 西太平洋 18.464 1°N 113.998 1°E 3 681 2015年6月30日 P20 西太平洋 18.563 9°N 113.674 3°E 3 743 2015年7月2日 注:P15–P17号样品,沉积物采集深度分别为0~2 cm、2~5 cm、5~10 cm;其余表层沉积物采集深度均为0~5 cm。 -
[1] Niu Mingyang, Liang Wenyue, Wang Fengping. Methane biotransformation in the ocean and its effects on climate change: a review[J]. Science China Earth Sciences, 2018, 61(12): 1697−1713. doi: 10.1007/s11430-017-9299-4 [2] Ferry J G, Lessner D J. Methanogenesis in marine sediments[J]. Annals of the New York Academy of Sciences, 2008, 1125(1): 147−157. doi: 10.1196/annals.1419.007 [3] Hartmann D L, Klein Tank A M G, Rusticucci M, et al. Climate Change 2013: the Physical Science Basis: Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change[M]. Cambridge: Cambridge University Press, 2014. [4] Lowe D C. Global change: a green source of surprise[J]. Nature, 2006, 439(7073): 148−149. doi: 10.1038/439148a [5] Niemann H, Lösekann T, de Beer D, et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink[J]. Nature, 2006, 443(7113): 854−858. doi: 10.1038/nature05227 [6] Giovannell D, d’Errico G, Fiorentino F, et al. Diversity and distribution of prokaryotes within a shallow-water pockmark field[J]. Frontiers in Microbiology, 2016, 7: 941. [7] Tu T H, Wu Liwei, Lin Y S, et al. Microbial community composition and functional capacity in a terrestrial ferruginous, sulfate-depleted mud volcano[J]. Frontiers in Microbiology, 2017, 8: 2137. doi: 10.3389/fmicb.2017.02137 [8] Jing Hongmei, Wang Ruonan, Jiang Qiuyun, et al. Anaerobic methane oxidation coupled to denitrification is an important potential methane sink in deep-sea cold seeps[J]. Science of the Total Environment, 2020, 748: 142459. doi: 10.1016/j.scitotenv.2020.142459 [9] Inagaki F, Kuypers M M M, Tsunogai U, et al. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system[J]. Proceedings of the National Academy of Sciences, 2006, 103(38): 14164−14169. doi: 10.1073/pnas.0606083103 [10] Boetius A, Wenzhöfer F. Seafloor oxygen consumption fuelled by methane from cold seeps[J]. Nature Geoscience, 2013, 6(9): 725−734. doi: 10.1038/ngeo1926 [11] D’Hondt S, Rutherford S, Spivack A J. Metabolic activity of subsurface life in deep-sea sediments[J]. Science, 2002, 295(5562): 2067−2070. doi: 10.1126/science.1064878 [12] Chen Jinquan, Wang Fengping, Zheng Yanping, et al. Investigation of the methanogen-related archaeal population structure in shallow sediments of the Pearl River Estuary, Southern China[J]. Journal of Basic Microbiology, 2014, 54(6): 482−490. doi: 10.1002/jobm.201200172 [13] Niu Mingyang, Fan Xibei, Zhuang Guangchao, et al. Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea[J]. FEMS Microbiology Ecology, 2017, 93(9): fix101. [14] Yang Shanshan, Lü Yongxin, Liu Xipeng, et al. Genomic and enzymatic evidence of acetogenesis by anaerobic methanotrophic archaea[J]. Nature Communications, 2020, 11(1): 3941. doi: 10.1038/s41467-020-17860-8 [15] Chen Ye, Li Siqi, Xu Xiaoqing, et al. Characterization of microbial communities in sediments of the South Yellow Sea[J]. Journal of Oceanology and Limnology, 2021, 39(3): 846−864. doi: 10.1007/s00343-020-0106-6 [16] Mihajlovski A, Alric M, Brugère J F. A putative new order of methanogenic archaea inhabiting the human gut, as revealed by molecular analyses of the mcrA gene[J]. Research in Microbiology, 2008, 159(7/8): 516−521. [17] Dridi B, Raoult D, Drancourt M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of Archaea: towards the universal identification of living organisms[J]. Acta Pathologica Microbiologica et Immunologica Scandinavica, 2012, 120(2): 85−91. doi: 10.1111/j.1600-0463.2011.02833.x [18] Wise M G, McArthur J V, Shimkets L J. Methanotroph diversity in landfill soil: isolation of novel type I and type II methanotrophs whose presence was suggested by culture-independent 16S ribosomal DNA analysis[J]. Applied and Environmental Microbiology, 1999, 65(11): 4887−4897. doi: 10.1128/AEM.65.11.4887-4897.1999 [19] Raghoebarsing A A, Pol A, van de Pas-Schoonen K T, et al. A microbial consortium couples anaerobic methane oxidation to denitrification[J]. Nature, 2006, 440(7086): 918−921. doi: 10.1038/nature04617 [20] Dhillon A, Lever M, Lloyd K G, et al. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin[J]. Applied and Environmental Microbiology, 2005, 71(8): 4592−4601. doi: 10.1128/AEM.71.8.4592-4601.2005 [21] 范习贝, 梁前勇, 牛明杨, 等. 中国南海北部陆坡沉积物古菌多样性及丰度分析[J]. 微生物学通报, 2017, 44(7): 1589−1601. doi: 10.13344/j.microbiol.china.170159Fan Xibei, Liang Qianyong, Niu Mingyang, et al. The diversity and richness of archaea in the northern continental slope of South China Sea[J]. Microbiology China, 2017, 44(7): 1589−1601. doi: 10.13344/j.microbiol.china.170159 [22] Prouty N G, Campbell P L, Close H G, et al. Molecular indicators of methane metabolisms at cold seeps along the United States Atlantic Margin[J]. Chemical Geology, 2020, 543: 119603. doi: 10.1016/j.chemgeo.2020.119603 [23] Santoro A E, Dupont C L, Richter R A, et al. Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: an ammonia-oxidizing archaeon from the open ocean[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(4): 1173−1178. doi: 10.1073/pnas.1416223112 [24] Hammer Ø, Harper D A, Ryan P D. PAST: paleontological statistics software package for education and data analysis[J]. Palaeontologia Electronica, 2001, 4(1): 1−9. [25] Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Molecular Biology and Evolution, 2016, 33(7): 1870−1874. doi: 10.1093/molbev/msw054 [26] 郭文捷. 热带西太平洋深海冷泉−海山极端环境微生物多样性研究[D]. 青岛: 青岛大学, 2017.Guo Wenjie. Microbial community diversity of the deep sea cold seep and seamount-extreme environments in tropical western Pacific[D]. Qingdao: Qingdao University, 2017. [27] Orphan V J, Jahnke L L, Embaye T, et al. Characterization and spatial distribution of methanogens and methanogenic biosignatures in hypersaline microbial mats of Baja California[J]. Geobiology, 2008, 6(4): 376−393. doi: 10.1111/j.1472-4669.2008.00166.x [28] Vigneron A, L’Haridon S, Godfroy A, et al. Evidence of active methanogen communities in shallow sediments of the Sonora Margin cold seeps[J]. Applied and Environmental Microbiology, 2015, 81(10): 3451−3459. doi: 10.1128/AEM.00147-15 [29] Postec A, Quéméneur M, Bes M, et al. Microbial diversity in a submarine carbonate edifice from the serpentinizing hydrothermal system of the Prony Bay (New Caledonia) over a 6-year period[J]. Frontiers in Microbiology, 2015, 6: 857. [30] Zhou Zhichao, Chen Jing, Cao Huiluo, et al. Analysis of methane-producing and metabolizing archaeal and bacterial communities in sediments of the northern South China Sea and coastal Mai Po Nature Reserve revealed by PCR amplification of mcrA and pmoA genes[J]. Frontiers in Microbiology, 2015, 5: 789. [31] 李涛, 王鹏, 汪品先. 南海南部陆坡表层沉积物细菌和古菌多样性[J]. 微生物学报, 2008, 48(3): 323−329. doi: 10.3321/j.issn:0001-6209.2008.03.009Li Tao, Wang Peng, Wang Pinxian. Bacterial and archaeal diversity in surface sediment from the south slope of the South China Sea[J]. Acta Microbiologica Sinica, 2008, 48(3): 323−329. doi: 10.3321/j.issn:0001-6209.2008.03.009 [32] 王风平, 周悦恒, 张新旭, 等. 深海微生物多样性[J]. 生物多样性, 2013, 21(4): 445−455.Wang Fengping, Zhou Yueheng, Zhang Xinxu, et al. Biodiversity of deep-sea microorganisms[J]. Biodiversity Science, 2013, 21(4): 445−455. [33] Claypool G E, Kvenvolden K A. Methane and other hydrocarbon gases in marine sediment[J]. Annual Review of Earth and Planetary Sciences, 1983, 11(1): 299−327. doi: 10.1146/annurev.ea.11.050183.001503 [34] Danovaro R, Molari M, Corinaldesi C, et al. Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems[J]. Science Advances, 2016, 2(4): e1500961. doi: 10.1126/sciadv.1500961 [35] Jing Hongmei, Xia Xiaomin, Liu Hongbin, et al. Anthropogenic impact on diazotrophic diversity in the mangrove rhizosphere revealed by nifH pyrosequencing[J]. Frontiers in Microbiology, 2015, 6: 1172. [36] Nunoura T, Nishizawa M, Kikuchi T, et al. Molecular biological and isotopic biogeochemical prognoses of the nitrification-driven dynamic microbial nitrogen cycle in hadopelagic sediments[J]. Environmental Microbiology, 2013, 15(11): 3087−3107. [37] Nunoura T, Takaki Y, Kazama H, et al. Microbial diversity in deep-sea methane seep sediments presented by SSU rRNA gene tag sequencing[J]. Microbes and Environments, 2012, 27(4): 382−390. doi: 10.1264/jsme2.ME12032 [38] Yanagawa K, Sunamura M, Lever M A, et al. Niche separation of methanotrophic archaea (ANME-1 and -2) in methane-seep sediments of the eastern Japan Sea offshore Joetsu[J]. Geomicrobiology Journal, 2011, 28(2): 118−129. doi: 10.1080/01490451003709334 [39] Nunoura T, Oida H, Nakaseama M, et al. Archaeal Diversity and distribution along thermal and geochemical gradients in hydrothermal sediments at the Yonaguni Knoll IV hydrothermal field in the southern Okinawa Trough[J]. Applied and Environmental Microbiology, 2010, 76(4): 1198−1211. doi: 10.1128/AEM.00924-09 [40] Schippers A, Kock D, Höft C, et al. Quantification of microbial communities in subsurface marine sediments of the Black Sea and off Namibia[J]. Frontiers in Microbiology, 2012, 3: 16. -





 下载:
下载: