Comparison of the composition and functional potentials of bacterial communities in different tissues from Crassostrea sikamea, Crassostrea angulata and Crassostrea gigas
-
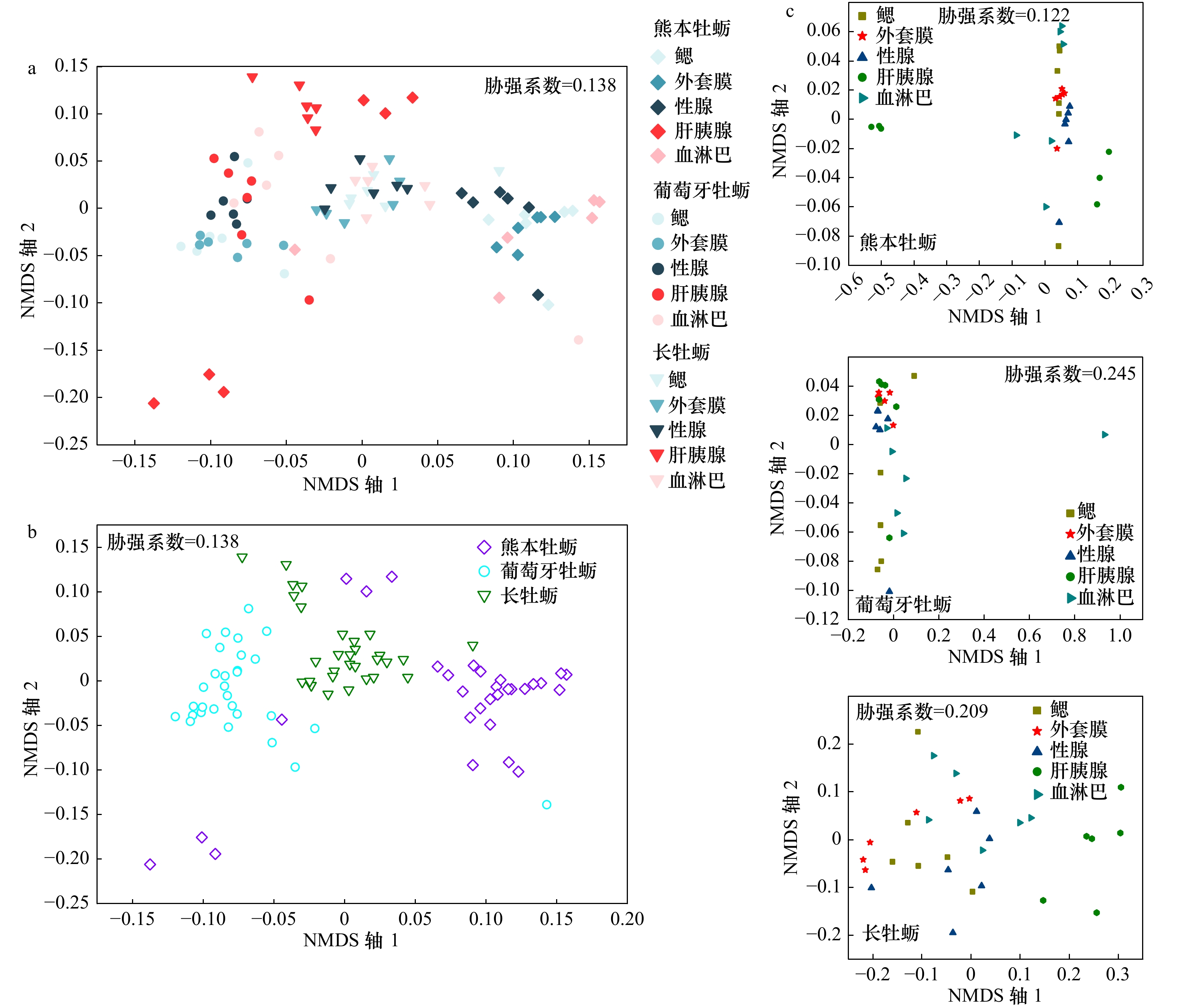
摘要: 为探究健康二龄熊本牡蛎、葡萄牙牡蛎和长牡蛎5种组织间细菌群落组成、多样性和功能的差异,本研究利用Illumina高通量测序技术和PICRUSt2对3种牡蛎5种组织中的细菌群落构成及其潜在功能进行了比较分析。结果显示,在3种牡蛎的5种组织中共鉴定到6 020个细菌OTUs,其中3种牡蛎整体间共有的OTUs数占42.4%。3种牡蛎5种组织中的优势菌主要隶属于γ-变形菌纲(γ-proteobacteria)、α-变形菌纲(α-proteobacteria)、柔壁菌门(Tenericutes)和厚壁菌门(Firmicutes)。弧菌属(Vibrio)在熊本牡蛎的鳃、外套膜、性腺和血淋巴中的相对丰度均显著高于长牡蛎和葡萄牙牡蛎,且分别在熊本牡蛎外套膜、长牡蛎性腺和葡萄牙牡蛎血淋巴中相对丰度最高,但均在3种牡蛎肝胰腺中相对丰度最低。3种牡蛎5种组织间的菌群多样性均有所差异,其中在熊本牡蛎鳃、葡萄牙牡蛎外套膜和长牡蛎血淋巴中菌群多样性最高,而在熊本牡蛎血淋巴、葡萄牙牡蛎性腺和长牡蛎肝胰腺中菌群多样性最低。在不区分组织的情况下,3种牡蛎整体间的菌群结构存在显著差异(r=0.661,p<0.001)。此外,在熊本牡蛎肝胰腺、葡萄牙牡蛎鳃和长牡蛎血淋巴中细菌参与的能量代谢相关功能通路的相关丰度显著高于对应牡蛎的其他组织。本文结果表明,熊本牡蛎、葡萄牙牡蛎和长牡蛎组织内的菌群构成存在差异,且由细菌介导的功能也随牡蛎种类和组织类型发生改变。Abstract: To explore the differences in the composition, diversity, and functions of bacterial communities in tissues from two-year old and healthy Crassostrea sikamea, Crassostrea angulata and Crassostrea gigas, the bacterial community structures and functional potentials among three oysters by combining the Illumina MiSeq high-throughput sequencing technology and phylogenetic investigation of communities by reconstruction of unobserved states 2 (PICRUSt2) was compared in this paper. The results showed that a total of 6 020 OTUs were identified in the five tissues from the three oysters, among which the numbers of shared OTUs accounted for 42.4% of total OTUs. The γ-proteobacteria, α-proteobacteria, Tenericutes and Firmicutes were dominant in five tissues of three oysters. The relative abundance of Vibrio genus was significantly higher in the gill, mantle, hepatopancreas and hemolymph tissues of C. sikamea than in the five tissues of C. angulata and C. gigas. Compared to other tissues, the relative abundance of Vibrio genus was the highest in the mantle of C. sikamea, the gonad of C. angulata and the hemolymph of C. gigas, but was lowest in the hepatopancreas of all three oysters. The diversity of bacterial communities was different among the five tissues of three oysters. Compared to other tissues, the bacterial community α-diversity was higher in the gill of C. sikamea, the mantle of C. angulata and the hemolymph of C. gigas, respectively, but was lowest in the hemolymph of C. sikamea, the gonad of C. angulata and the hepatopancreas of C. gigas. Regardless of tissue types, the bacterial community structures differed significantly (r=0.661, p<0.001) among the three oysters. Additionally, the abundances of bacterial-mediated functional pathways involved in the energy metabolism were significantly higher in the hepatopancreas of C. sikamea, the gill of C. angulata and the hemolymph of C. gigas than that in other tissues of corresponding oysters. Our findings suggested that the structure and composition of bacterial communities were different among five tissues of C. sikamea, C. angulata and C. gigas, and the bacterial-mediated functional potentials ware affected by oyster species and tissue types.
-
Key words:
- oyster /
- high-throughput sequencing /
- bacterial community /
- function prediction
-
图 3 3种牡蛎5种组织中的优势细菌门(变形菌门归类到纲)(a)和细菌属(b)的平均相对丰度
Cs. 熊本牡蛎;Ca. 葡萄牙牡蛎;Cg. 长牡蛎
Fig. 3 Average relative abundances of the dominant bacterial phyla (Proteobacteria were assigned to the class level) (a) and genus (b) in the five tissues form three oysters
Cs. Crassostrea sikamea; Ca. Crassostrea angulate; Cg. Crassostrea gigas
图 4 比较3种牡蛎5种组织中的细菌群落α-多样性差异
Cs. 熊本牡蛎;Ca. 葡萄牙牡蛎;Cg. 长牡蛎;采用单因素方差分析3种牡蛎种类间在相同组织情况下细菌群落α-多样性的差异,不同字母表示3种牡蛎种类具有差异性( p<0.05)
Fig. 4 Comparison of bacterial community α-diversity in five tissues from three oysters
Cs. Crassostrea sikamea; Ca. Crassostrea angulate; Cg. Crassostrea gigas; the difference of bacterial community α-diversity among three oysters under the same tissue was analyzed by one-way variance analysis, of which showed differences using the different letters (p<0.05)
图 6 热图展示3种牡蛎的肝胰腺、鳃、血淋巴中细菌群落功能途径的丰度(平方根转换)分布
Cs. 熊本牡蛎;Ca. 葡萄牙牡蛎;Cg. 长牡蛎
Fig. 6 Heatmap showing the abundance (sqrt (x) transformed) distribution of functional pathways of bacterial communities in the gill, hepatopancreas, and hemolymph from three oysters
Cs. Crassostrea sikamea; Ca. Crassostrea angulate; Cg. Crassostrea gigas
表 1 基于Bray-Curtis距离比较3种牡蛎不同组织间细菌群落的整体差异性
Tab. 1 Comparison of overall differences in bacterial communities among different tissues of the three oysters based on Bray-Curtis distance
组别 所有组织 鳃 肝胰腺 外套膜 性腺 血淋巴 R p R p R p R p R p R p 3种牡蛎 0.661 0.001 0.721 0.001 0.580 0.001 0.979 0.001 0.915 0.001 0.478 0.001 Cs vs Ca 0.633 0.001 1.000 0.003 0.283 0.062 1.000 0.003 0.998 0.003 0.437 0.014 Cs vs Cg 0.486 0.001 0.724 0.003 0.435 0.003 1.000 0.003 0.909 0.003 0.543 0.009 Ca vs Cg 0.532 0.001 0.769 0.003 0.885 0.003 0.800 0.003 0.874 0.003 0.572 0.003 注:加粗的数值表示差异显著(p<0.05)。 表 2 基于 Bray-Curtis 距离比较同种牡蛎不同组织间细菌群落的差异性
Tab. 2 Comparison of difference in bacterial communities among different tissues from the same oyster based on Bray-Curtis distance
牡蛎种类 组织类型 鳃 外套膜 性腺 肝胰腺 血淋巴 熊本牡蛎 鳃 0.247 0.465 0.471 0.013 外套膜 0.014 0.426 0.441 0.148 性腺 0.003 0.011 0.501 0.38 肝胰腺 0.003 0.002 0.002 0.462 血淋巴 0.312 0.071 0.002 0.011 葡萄牙牡蛎 鳃 0.482 0.057 0.357 0.283 外套膜 0.004 0.194 0.244 0.445 性腺 0.220 0.028 0.217 0.383 肝胰腺 0.025 0.030 0.034 0.4 血淋巴 0.005 0.005 0.004 0.010 长牡蛎 鳃 0.413 0.107 0.948 0.53 外套膜 0.002 0.507 1.000 0.625 性腺 0.096 0.005 0.967 0.369 肝胰腺 0.003 0.002 0.002 0.957 血淋巴 0.002 0.002 0.007 0.003 注:每个种类的上三角和下三角分别为样本间细菌群落差异性的R值和p值;加粗的数值表示差异显著(p<0.05)。 表 3 基于非参数的多元置换方差分析牡蛎种类和组织类型对细菌群落的定量影响
Tab. 3 Quantitative effects of oyster species and tissue types on the variations in bacterial community based on nonparametric permutational multivariate analysis of variance (PERMANOVA)
自由度 平方和 均方差 F模型 R2 p 牡蛎种类 2 6.888 3.444 20.283 0.258 0.001 组织类型 12 7.117 0.593 3.493 0.266 0.001 残差 75 12.735 0.170 − 0.476 − 总计 89 26.741 − − 1 − 注:加粗的数值表示差异显著(p<0.05);−代表无数值。 -
[1] Clerissi C, de Lorgeril J, Petton B, et al. Diversity and stability of microbiota are key factors associated to healthy and diseased Crassostrea gigas oysters[J]. BioRxiv, 2018, doi: 10.1101/378125. [2] Wang Yanting, Wang Kai, Huang Lei, et al. Fine-scale succession patterns and assembly mechanisms of bacterial community of Litopenaeus vannamei larvae across the developmental cycle[J]. Microbiome, 2020, 8(1): 106. doi: 10.1186/s40168-020-00879-w [3] Semova I, Carten J D, Stombaugh J, et al. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish[J]. Cell Host & Microbe, 2012, 12(3): 277−288. [4] Modak T H, Gomez-Chiarri M. Contrasting immunomodulatory effects of probiotic and pathogenic bacteria on eastern oyster, Crassostrea virginica, larvae[J]. Vaccines, 2020, 8(4): 588. doi: 10.3390/vaccines8040588 [5] Kesarcodi-Watson A, Miner P, Nicolas J L, et al. Protective effect of four potential probiotics against pathogen-challenge of the larvae of three bivalves: Pacific oyster (Crassostrea gigas), flat oyster (Ostrea edulis) and scallop (Pecten maximus)[J]. Aquaculture, 2012, 344−349: 29−34. doi: 10.1016/j.aquaculture.2012.02.029 [6] 全为民, 张锦平, 平仙隐, 等. 巨牡蛎对长江口环境的净化功能及其生态服务价值[J]. 应用生态学报, 2007, 18(4): 871−876. doi: 10.3321/j.issn:1001-9332.2007.04.028Quan Weimin, Zhang Jinping, Ping Xianyin, et al. Purification function and ecological services value of Crassostrea sp. in Yangtze River Estuary[J]. Chinese Journal of Applied Ecology, 2007, 18(4): 871−876. doi: 10.3321/j.issn:1001-9332.2007.04.028 [7] Lokmer A, Kuenzel S, Baines J F, et al. The role of tissue-specific microbiota in initial establishment success of Pacific oysters[J]. Environmental Microbiology, 2016, 18(3): 970−987. doi: 10.1111/1462-2920.13163 [8] Sohn S, Lundgren K M, Tammi K, et al. Probiotic strains for disease management in hatchery larviculture of the eastern oyster Crassostrea virginica[J]. Journal of Shellfish Research, 2016, 35(2): 307−317. doi: 10.2983/035.035.0205 [9] Ruiz-Ponte C, Samain J F, Sánchez J L, et al. The benefit of a roseobacter species on the survival of scallop larvae[J]. Marine Biotechnology, 1999, 1(1): 52−59. doi: 10.1007/PL00011751 [10] Stevick R J, Sohn S, Modak T H, et al. Bacterial community dynamics in an oyster hatchery in response to probiotic treatment[J]. Frontiers in Microbiology, 2019, 10: 1060. doi: 10.3389/fmicb.2019.01060 [11] Dubert J, Barja J L, Romalde J L. New insights into pathogenic vibrios affecting bivalves in hatcheries: present and future prospects[J]. Frontiers in Microbiology, 2017, 8: 762. doi: 10.3389/fmicb.2017.00762 [12] 江海洋, 李磊, 莫宝庆, 等. 贝类污染副溶血性弧菌与海水水质相关性研究[J]. 中国卫生检验杂志, 2008, 18(12): 2502−2504. doi: 10.3969/j.issn.1004-8685.2008.12.018Jiang Haiyang, Li Lei, Mo Baoqing, et al. Relationship between Vibrio parahaemolyticus concentration in shellfish and sea water quality[J]. Chinese Journal of Health Laboratory Technology, 2008, 18(12): 2502−2504. doi: 10.3969/j.issn.1004-8685.2008.12.018 [13] Larsen A M, Mohammed H H, Arias C R. Characterization of the gut microbiota of three commercially valuable warmwater fish species[J]. Journal of Applied Microbiology, 2014, 116(6): 1396−1404. doi: 10.1111/jam.12475 [14] Zhang Xuechen, Li Xiaohui, Lu Jiaqi, et al. Quantifying the importance of external and internal sources to the gut microbiota in juvenile and adult shrimp[J]. Aquaculture, 2021, 531: 735910. doi: 10.1016/j.aquaculture.2020.735910 [15] 孙雪莹. 虾夷扇贝幼体及育苗池水体细菌群落动态及潜在益生菌筛选[D]. 大连: 大连海洋大学, 2016.Sun Xueying. Dynamics of bacterial communities associated with yesso scallop (Patinopecten yessoensisis) larvae and in the water of larval-rearing tanks, and selection of potential probiotics[D]. Dalian: Dalian Ocean University, 2016. [16] 陈琼, 李贵阳, 罗坤, 等. 凡纳滨对虾(Litopenaeus vannamei)亲虾繁殖期水体微生物多样性[J]. 海洋与湖沼, 2017, 48(1): 130−138.Chen Qiong, Li Guiyang, Luo Kun, et al. Microbial diversity in broodstock waters of the two genders of Litopenaeus vannamei[J]. Oceanologia et Limnologia Sinica, 2017, 48(1): 130−138. [17] Zhang Zhen, Lu Zhimeng, Zhang Weiwei, et al. Comparative analysis of midgut bacterial community under Vibrio splendidus infection in Apostichopus japonicus with hindgut as a reference[J]. Aquaculture, 2019, 513: 734427. doi: 10.1016/j.aquaculture.2019.734427 [18] 郁维娜, 戴文芳, 陶震, 等. 健康与患病凡纳滨对虾肠道菌群结构及功能差异研究[J]. 水产学报, 2018, 42(3): 399−409.Yu Weina, Dai Wenfang, Tao Zhen, et al. Characterizing the compositional and functional structures of intestinal microflora between healthy and diseased Litopenaeus vannamei[J]. Journal of Fisheries of China, 2018, 42(3): 399−409. [19] Wei Yongjun, Ren Tianqi, Zhang Lei. Dix-seq: an integrated pipeline for fast amplicon data analysis[J]. BioRxiv, 2020, doi: 10.1101/2020.05.11.089748. [20] Magoč T, Salzberg S L. FLASH: fast length adjustment of short reads to improve genome assemblies[J]. Bioinformatics, 2011, 27(21): 2957−2963. doi: 10.1093/bioinformatics/btr507 [21] Edgar R C. Search and clustering orders of magnitude faster than BLAST[J]. Bioinformatics, 2010, 26(19): 2460−2461. doi: 10.1093/bioinformatics/btq461 [22] Caporaso J G, Bittinger K, Bushman F D, et al. PyNAST: a flexible tool for aligning sequences to a template alignment[J]. Bioinformatics, 2010, 26(2): 266−267. doi: 10.1093/bioinformatics/btp636 [23] Pierce M L, Ward J E. Gut microbiomes of the eastern oyster (Crassostrea virginica) and the blue mussel (Mytilus edulis): temporal variation and the influence of marine aggregate-associated microbial communities[J]. mSphere, 2019, 4(6): e00730−19. [24] 王鑫毅, 谢骁, 金珊, 等. 基于高通量测序的缢蛏及其养殖池塘菌群结构的季节变化[J]. 应用生态学报, 2019, 30(12): 4267−4276.Wang Xinyi, Xie Xiao, Jin Shan, et al. Seasonal variation of microflora in Sinonovacula constricta and its aquacultural pond based on high-throughput sequencing[J]. Chinese Journal of Applied Ecology, 2019, 30(12): 4267−4276. [25] Pimentel Z T, Dufault-Thompson K, Russo K T, et al. Microbiome analysis reveals diversity and function of mollicutes associated with the eastern oyster, Crassostrea virginica[J]. mSphere, 2021, 6(3): e00227−21. [26] Tanaka R, Ootsubo M, Sawabe T, et al. Biodiversity and in situ abundance of gut microflora of abalone (Haliotis discus hannai) determined by culture-independent techniques[J]. Aquaculture, 2004, 241(1/4): 453−463. [27] 徐嘉康, 王劲松, 方怡涵, 等. 厚壳贻贝肠道细菌的生物被膜对其幼虫和稚贝附着的影响[J]. 海洋学报, 2021, 43(9): 81−91.Xu Jiakang, Wang Jinsong, Fang Yihan, et al. Effects of intestinal bacterial biofilms on settlement process of larvae and plantigrades in Mytilus coruscus[J]. Haiyang Xuebao, 2021, 43(9): 81−91. [28] Karimi E, Keller-Costa T, Slaby B M, et al. Genomic blueprints of sponge-prokaryote symbiosis are shared by low abundant and cultivatable Alphaproteobacteria[J]. Scientific Reports, 2019, 9(1): 1999. doi: 10.1038/s41598-019-38737-x [29] 李玲玲, 谢超伊, 宋宏策, 等. 长牡蛎(Crassostrea gigas)黑色壳表面微生物多样性的研究[J]. 海洋与湖沼, 2021, 52(6): 1418−1429. doi: 10.11693/hyhz20210400082Li Lingling, Xie Chaoyi, Song Hongce, et al. Microbial diversity on surface of black-shell Crassostrea gigas[J]. Oceanologia et Limnologia Sinica, 2021, 52(6): 1418−1429. doi: 10.11693/hyhz20210400082 [30] 邹建威. 岩扇贝内脏团和肠道可培养微生物及其宏基因多样性分析[D]. 大连: 大连海洋大学, 2019.Zou Jianwei. Diversity analysis of culturable microorganisms and their macrogenes in rock scallop viscera and intestine[D]. Dalian: Dalian Ocean University, 2019. [31] Wang Hailiang, Sun Bochao, Xie Guosi, et al. Spotlight on a novel bactericidal mechanism and a novel SXT/R391-like integrative and conjugative element, carrying multiple antibiotic resistance genes, in Pseudoalteromonas flavipulchra strain CDM8[J]. Microbiological Research, 2021, 242: 126598. doi: 10.1016/j.micres.2020.126598 [32] Bairagi A, Ghosh K S, Sen S K, et al. Evaluation of the nutritive value of Leucaena leucocephala leaf meal, inoculated with fish intestinal bacteria Bacillus subtilis and Bacillus circulans in formulated diets for rohu, Labeo rohita (Hamilton) fingerlings[J]. Aquaculture Research, 2004, 35(5): 436−446. doi: 10.1111/j.1365-2109.2004.01028.x [33] Dai Wenfang, Dong Yinghui, Ye Jing, et al. Gut microbiome composition likely affects the growth of razor clam Sinonovacula constricta[J]. Aquaculture, 2022, 550: 737847. doi: 10.1016/j.aquaculture.2021.737847 [34] Gradoville M R, Crump B C, Häse C C, et al. Environmental controls of oyster-pathogenic Vibrio spp. in oregon estuaries and a shellfish hatchery[J]. Applied and Environmental Microbiology, 2018, 84(9): e02156−17. [35] Beaz-Hidalgo R, Balboa S, Romalde J L, et al. Diversity and pathogenecity of Vibrio species in cultured bivalve molluscs[J]. Environmental Microbiology Reports, 2010, 2(1): 34−43. doi: 10.1111/j.1758-2229.2010.00135.x [36] Le Roux F, Wegner K M, Polz M F. Oysters and vibrios as a model for disease dynamics in wild animals[J]. Trends in Microbiology, 2016, 24(7): 568−580. doi: 10.1016/j.tim.2016.03.006 [37] 郑国兴, 李何, 黄宁宇, 等. 文蛤病原菌(溶藻弧菌)的分离与性状及病文蛤组织的电镜观察[J]. 水产学报, 1991, 15(2): 85−95.Zheng Guoxing, Li He, Huang Ningyu, et al. Characteristics of Vibrio alginolyticus isolated from diseased clam Meretrix meretrix and histopathological observations on diseased clam by electron microscope[J]. Journal of Fisheries of China, 1991, 15(2): 85−95. [38] 高晓建, 姚东瑞, 孙晶晶, 等. 4株长牡蛎(Crassostrea gigas)致病性哈维氏弧菌(Vibrio harveyi)鉴定及其毒力基因检测[J]. 海洋湖沼通报, 2015(3): 87−96.Gao Xiaojian, Yao Dongrui, Sun Jingjing, et al. Identification of 4 pathogenic Vibrio harveyi strains isolated from diseased oyster (Crassostrea gigas) and detection of their virulence genes[J]. Transactions of Oceanology and Limnology, 2015(3): 87−96. [39] 胡慧雯. 浙江省贝类海产品副溶血弧菌污染的风险识别与评估[D]. 杭州: 浙江大学, 2017.Hu Huiwen. Risk identification and assessment of Vibro parahaemolyticus in shellfish seafood in Zhejiang[D]. Hangzhou: Zhejiang University, 2017. [40] Arfken A, Song B, Allen Jr S K, et al. Comparing larval microbiomes of the eastern oyster (Crassostrea virginica) raised in different hatcheries[J]. Aquaculture, 2021, 531: 735955. doi: 10.1016/j.aquaculture.2020.735955 [41] 柴英辉, 高菲, 王金锋, 等. 仿刺参(Apostichopus japonicus)肠道菌群的地域性差异与共性研究[J]. 海洋与湖沼, 2019, 50(5): 1127−1137. doi: 10.11693/hyhz20190200044Chai Yinghui, Gao Fei, Wang Jinfeng, et al. Intestinal microbiota in Apostichopus japonicus: regional difference and common feature[J]. Oceanologia et Limnologia Sinica, 2019, 50(5): 1127−1137. doi: 10.11693/hyhz20190200044 [42] Fernández N T, Mazón-Suástegui J M, Vázquez-Juárez R, et al. Changes in the composition and diversity of the bacterial microbiota associated with oysters (Crassostrea corteziensis, Crassostrea gigas and Crassostrea sikamea) during commercial production[J]. FEMS Microbiology Ecology, 2014, 88(1): 69−83. doi: 10.1111/1574-6941.12270 [43] 张令帅. 海洋酸化对长牡蛎(Crassostrea gigas)肝胰腺生理功能和能量供给策略的影响[D]. 北京: 中国科学院海洋研究所, 2020.Zhang Lingshuai. Impact of ocean acidification on hepatopancreas physiological function and energy supply of Pacific oyster (Crassostrea gigas)[D]. Beijing: Institute of Oceanology, Chinese Academy of Sciences, 2020. -





 下载:
下载: