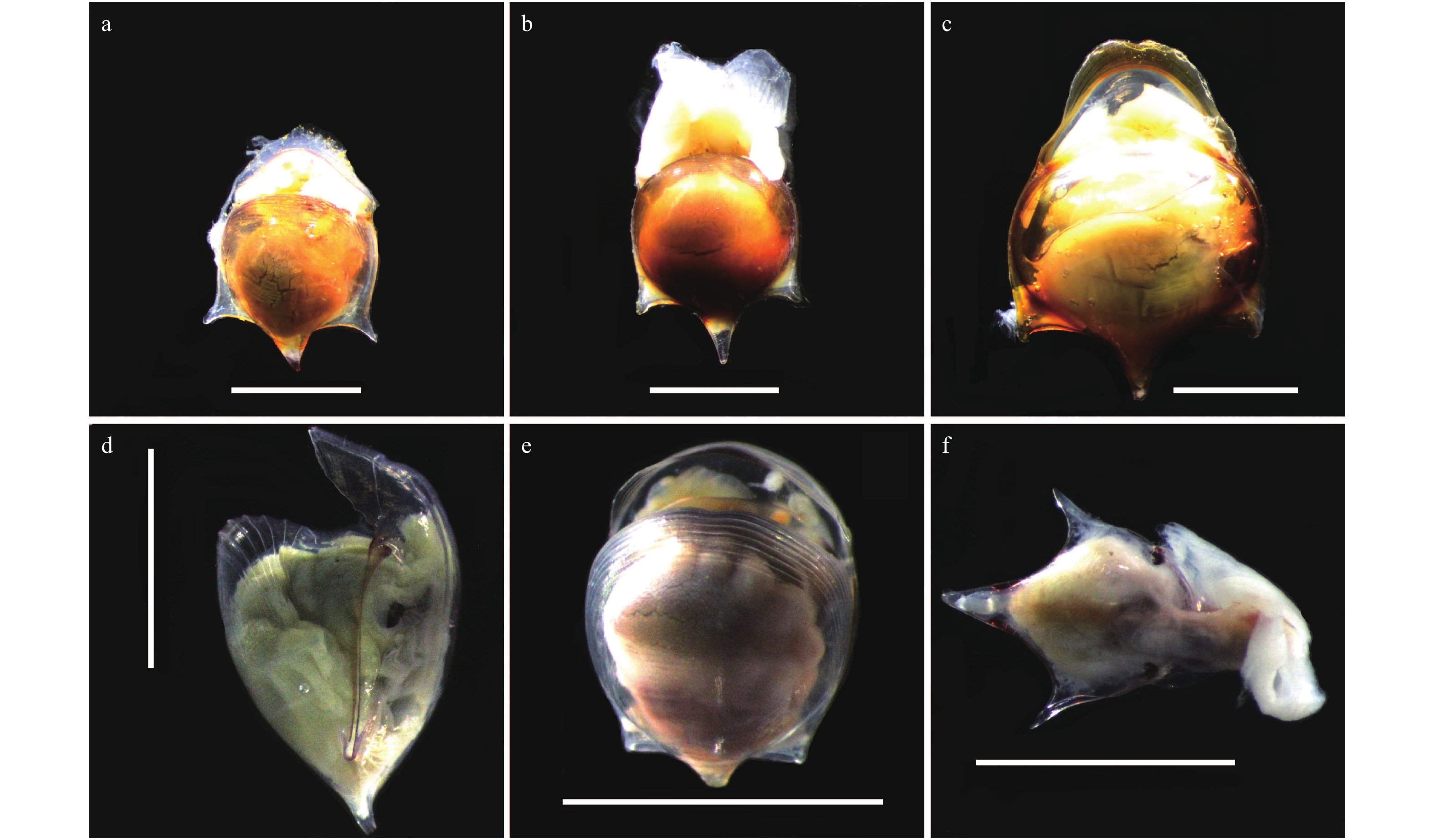
Taxonomy and DNA barcoding in genera Cavolinia and Diacavolinia from the Indian Ocean and Northwest Pacific Ocean
-
摘要: 近来的研究表明,一些所谓的环球或环极地分布的广布种实际上包含着一些局限性分布的隐存种,物种多样性可能被低估。本文采用形态学和DNA条形码技术相结合的方式,对印度洋和西北太平洋海域的龟螺属(Cavolinia)和小龟螺属(Diacavolinia)的种类进行了分类学研究和物种鉴定。结果表明,线粒体16S rRNA基因数据不支持小龟螺属形态种的划分,分布于西北太平洋的D. grayi、D. vanutrechti、D. pacifica、D. elegans、D. angulosa等多个形态种可能属同一个种,即长吻小龟螺(D. longirostris)。COI基因数据也不支持钩龟螺(C. uncinata)亚种和变形的划分。许多形态特征不能作为种或种下分类单元的区分依据。钩龟螺、球龟螺(C. globulosa)和长吻小龟螺在COI系统树中均形成2个地理支系,其内部可能存在隐存种。西北太平洋海域长吻小龟螺的核基因组中存在线粒体假基因,对DNA条形码分析产生严重干扰。Abstract: Some so-called circumglobal and/or circumpolar marine species are proved to be complexes of cryptic species with restricted distributions by molecular analyses. These phenomena imply that marine species diversity has been underestimated. In this study we use shell morphological characteristics together with mitochondrial COI and 16S rRNA gene sequences in order to study the taxonomy and to identify species of genera Cavolinia and Diacavolinia in family Cavoliniidae from the Indian Ocean and Northwest Pacific Ocean. Our results showed that the species delimitation of Diacavolinia is not supported by molecular analyses, those morphospecies (D. grayi, D. vanutrechti, D. pacifica, D. elegans, D. angulosa, etc.) with distinct morphology from northwestern Pacific are a single species, namely D. longirostris. The subdivision of subspecies and/or forms for C. uncinata could not be confirmed by DNA evidence. Phylogenetic analysis of COI revealed two independent geographical lineages for C. uncinata, C. globulosa and D. longirostris respectively, suggesting that these morphspecies may harbour cryptic diversity. Nuclear mitochondrial pseudogene sequences of COI were detected from most individuals of D. longirostris, which can affect the analysis of DNA barcoding.
-
Key words:
- Cavolinia /
- Diacavolinia /
- taxonomy /
- COI /
- 16S rRNA
-
图 4 基于16S rRNA基因构建的小龟螺和其他腹足类邻接系统发育树
节点处数值分别为自展支持率和后验概率,仅给出大于70%的数值;手绘图引自文献[25]
Fig. 4 Neighbor-joining phylogenetic tree of genus Diacavolinia and other gastropods based on mitochondrial 16S rRNA gene sequences
Bootstrap values/posterior probabilities over 70% are shown for each node. Drawings are reproduced from reference[25]
图 5 基于COI基因构建的龟螺属和小龟螺属贝叶斯系统发育树
节点处数值分别为后验概率和自展支持率,仅给出大于70%的数值;手绘图引自文献[24]
Fig. 5 Bayesian inference phylogenetic tree of genera Cavolinia and Diacavolinia based on mitochondrial COI gene sequences
Posterior probabilities/bootstrap values over 70% are shown for each node. Drawings are reproduced from reference [24]
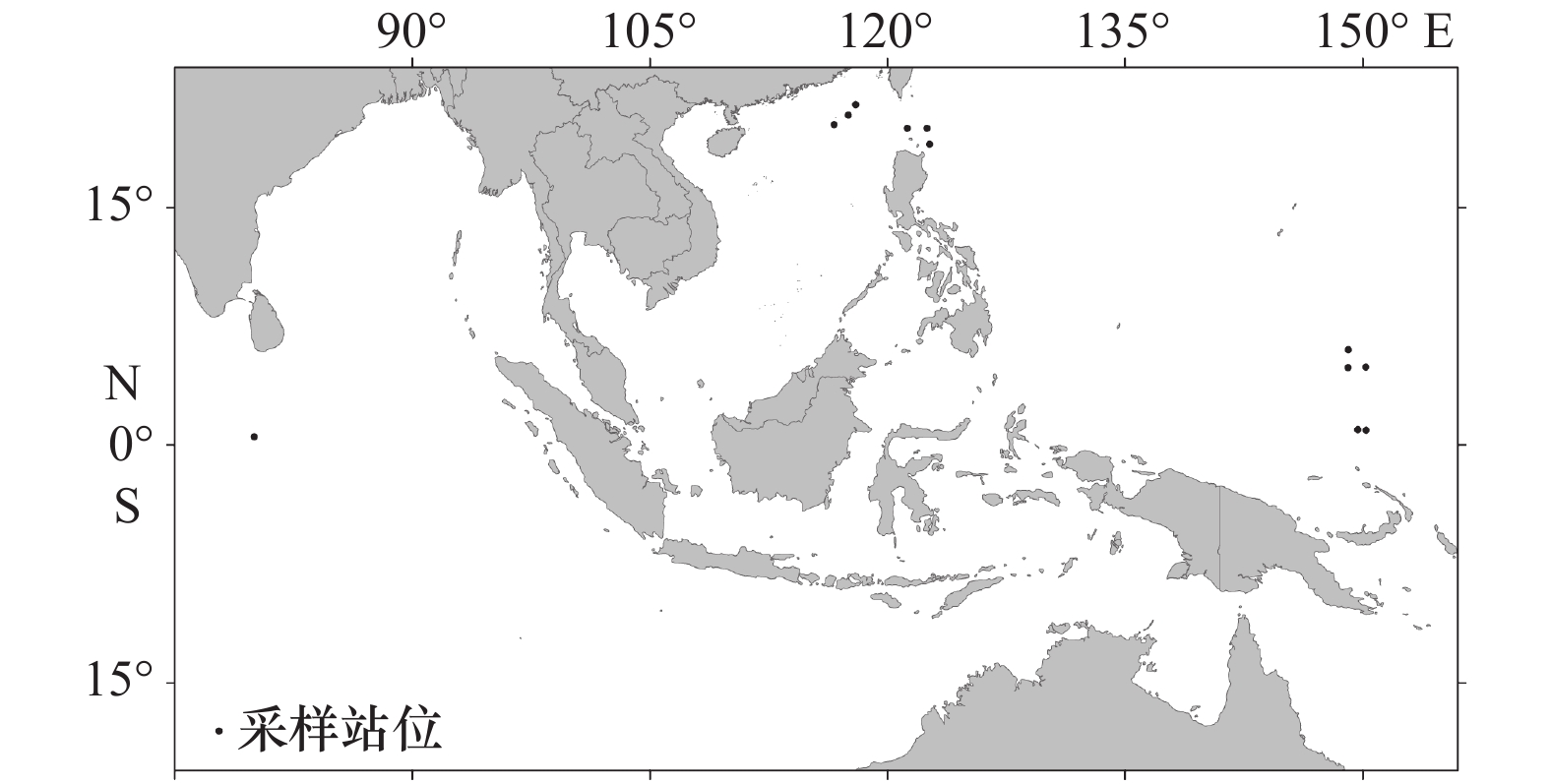
表 1 样品采集信息和基因库(GenBank)登录号
Tab. 1 Collection information and GenBank accession numbers for specimens analyzed in this study
形态种 测序标本数 采集海域 GenBank登录号 COI 16S rRNA Cavolinia uncinata 5 西北太平洋 MK913370~374 C. gibbosa 4 西北太平洋 MK913375~378 C. globulosa 5 西北太平洋、印度洋 MK913379~383 C. labiata 8 西北太平洋 MK913384~391 Diacavolinia vanutrechti 2 西北太平洋 MK913392 MK913407 Diacavolinia sp. 3 西北太平洋 MK913393~395 D. grayi 2 西北太平洋 MK913396/MK913399 D. angulosa 10 西北太平洋 MK913397~398/400~404/410~412 D. elegans 2 西北太平洋 MK913405~406 D. pacifica 2 西北太平洋 MK913408~409 -
[1] Doney S C, Fabry V J, Feely R A, et al. Ocean acidification: the other CO2 problem[J]. Annual Review of Marine Science, 2009, 1: 169−192. doi: 10.1146/annurev.marine.010908.163834 [2] Bednaršek N, Harvey C J, Kaplan I C, et al. Pteropods on the edge: cumulative effects of ocean acidification, warming, and deoxygenation[J]. Progress in Oceanography, 2016, 145: 1−24. doi: 10.1016/j.pocean.2016.04.002 [3] Alvarez-Fernandez S, Bach L T, Taucher J, et al. Plankton responses to ocean acidification: the role of nutrient limitation[J]. Progress in Oceanography, 2018, 165: 11−18. doi: 10.1016/j.pocean.2018.04.006 [4] Dupont S, Pörtner H. Get ready for ocean acidification[J]. Nature, 2013, 498(7455): 429. doi: 10.1038/498429a [5] Bunse C, Lundin D, Karlsson C M G, et al. Response of marine bacterioplankton pH homeostasis gene expression to elevated CO2[J]. Nature Climate Change, 2016, 6: 483−487. doi: 10.1038/nclimate2914 [6] Gardner J, Manno C, Bakker D C E, et al. Southern Ocean pteropods at risk from ocean warming and acidification[J]. Marine Biology, 2018, 165: 8. doi: 10.1007/s00227-017-3261-3 [7] Harley C D G, Hughes A R, Hultgren K M, et al. The impacts of climate change in coastal marine systems[J]. Ecology Letters, 2006, 9(2): 228−241. doi: 10.1111/j.1461-0248.2005.00871.x [8] Foster B A, Cargill J M, Montgomery J C. Planktivory in Pagothenia borchgrevinki (Pisces: Nototheniidae) in McMurdo Sound, Antarctica[J]. Polar Biology, 1987, 8(1): 49−54. doi: 10.1007/BF00297164 [9] Karnovsky N J, Hobson K A, Iverson S, et al. Seasonal changes in diets of seabirds in the North Water Polynya: a multiple-indicator approach[J]. Marine Ecology Progress Series, 2008, 357: 291−299. doi: 10.3354/meps07295 [10] Seibel B A, Dymowska A, Rosenthal J. Metabolic temperature compensation and coevolution of locomotory performance in pteropod molluscs[J]. Integrative and Comparative Biology, 2007, 47(6): 880−891. doi: 10.1093/icb/icm089 [11] Mucci A. The solubility of calcite and aragonite in seawater at various salinities, temperatures, and one atmosphere total pressure[J]. American Journal of Science, 1983, 283(7): 780−799. doi: 10.2475/ajs.283.7.780 [12] Roger L M, Richardson A J, McKinnon A D, et al. Comparison of the shell structure of two tropical Thecosomata (Creseis acicula and Diacavolinia longirostris) from 1963 to 2009: potential implications of declining aragonite saturation[J]. ICES Journal of Marine Science, 2012, 69(3): 465−474. doi: 10.1093/icesjms/fsr171 [13] Hunt B, Strugnell J, Bednarsek N, et al. Poles apart: the “bipolar” pteropod species Limacina helicina is genetically distinct between the Arctic and Antarctic oceans[J]. PLoS One, 2010, 5(3): e9835. doi: 10.1371/journal.pone.0009835 [14] 张福绥. 中国近海的浮游软体动物 I. 翼足类、异足类及海蜗牛类的分类研究[J]. 海洋科学集刊, 1964(5): 125−226.Zhang Fusui. The pelagic molluscs off China coast I. A systemic study of Pteropoda (Opisthobranchia), Heteropoda (Prosobranchia) and Janthinidae (Ptenoglossa, Prosobranchia)[J]. Studia Marina Sinica, 1964(5): 125−226. [15] 刘瑞玉. 中国海洋生物名录[M]. 北京: 科学出版社, 2008.Liu Ruiyu. Checklist of Marine Biota of China Seas[M]. Beijing: Science Press, 2008. [16] van der Spoel S. Euthecosomata, a Group with Remarkable Developmental Stages (Gastropoda, Pteropoda)[M]. Gorinchem: J. Noorduyn & ZN, 1967. [17] Rampal J. Les thécosomes (Mollusques pélagiques): systématique et évolution, écologie et biogéographie Méditerranéennes[D]. Marseille: Université de Provence, 1975. [18] Robertson R. Dispersal and wastage of larval Philippia krebsii (Gastropoda: Architectonicidae) in the north Atlantic[J]. Proceedings of the Academy of Natural Sciences of Philadelphia, 1964, 116: 1−27. [19] Bontes B, van der Spoel S. Variation in the Diacria trispinosa group, new interpretation of colour patterns and description of D. rubecula n. sp. (Pteropoda)[J]. Bulletin Zoölogisch Museum Universiteit van Amsterdam, 1998, 16(11): 77−84. [20] Burridge A K, Janssen A W, Peijnenburg K T C A. Revision of the genus Cuvierina Boas, 1886 based on integrative taxonomic data, including the description of a new species from the Pacific Ocean (Gastropoda, Thecosomata)[J]. ZooKeys, 2016(619): 1−12. [21] Hebert P D N, Cywinska A, Ball S L, et al. Biological identifications through DNA barcodes[J]. Proceedings of the Royal Society of London Series B: Biological Scences, 2003, 270(1512): 313−321. doi: 10.1098/rspb.2002.2218 [22] Jennings R M, Bucklin A, Ossenbrügger H, et al. Species diversity of planktonic gastropods (Pteropoda and Heteropoda) from six ocean regions based on DNA barcode analysis[J]. Deep Sea Research II: Topical Studies in Oceanography, 2010, 57(24/26): 2199−2210. [23] Gasca R, Janssen A W. Taxonomic review, molecular data and key to the species of Creseidae from the Atlantic Ocean[J]. Journal of Molluscan Studies, 2014, 80(1): 35−42. doi: 10.1093/mollus/eyt038 [24] van der Spoel S. New forms of Diacria quadridentata (de Blainville, 1821), Cavolinia longirostris (de Blainville, 1821) and Cavolinia uncinata (Rang, 1829) from the Red Sea and the East Pacific Ocean (Mollusca, Pteropoda)[J]. Beaufortia, 1971, 19(243): 1−20. [25] van der Spoel S, Bleeker J, Kobayasi H. From Cavolinia longirostris to twenty-four Diacavolinia taxa, with a phylogenetic discussion (Mollusca, Gastropoda)[J]. Bijdragen tot de Dierkunde, 1993, 62(3): 127−166. doi: 10.1163/26660644-06203001 [26] Tesch J J. The thecosomatous pteropods. I. The Atlantic[J]. Dana Report, 1946, 28: 1−82. [27] Tesch J J. The thecosomatous pteropods II. The Indo-Pacific[J]. Dana Report, 1948, 30: 1−45. [28] Maas A E, Blanco-Bercial L, Lawson G L. Reexamination of the species assignment of Diacavolinia pteropods using DNA barcoding[J]. PLoS One, 2013, 8(1): e53889. doi: 10.1371/journal.pone.0053889 [29] 李海涛, 何薇, 周鹏, 等. 伶鼬榧螺(Oliva mustelina)的分子鉴定及其形态变异[J]. 海洋学报, 2015, 37(4): 117−123.Li Haitao, He Wei, Zhou Peng, et al. Molecular identification of Oliva mustelina and its morphological variation[J]. Haiyang Xuebao, 2015, 37(4): 117−123. [30] Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates[J]. Molecular Marine Biology and Biotechnology, 1994, 3(5): 294−299. [31] Simon C, Frati F, Beckenbach A, et al. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers[J]. Annals of the Entomological Society of America, 1994, 87(6): 651−701. doi: 10.1093/aesa/87.6.651 [32] Xia Xuhua. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution[J]. Molecular Biology and Evolution, 2013, 30(7): 1720−1728. doi: 10.1093/molbev/mst064 [33] Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Molecular Biology and Evolution, 2016, 33(7): 1870−1874. doi: 10.1093/molbev/msw054 [34] Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space[J]. Systematic Biology, 2012, 61(3): 539−542. doi: 10.1093/sysbio/sys029 [35] Darriba D, Taboada G L, Doallo R, et al. jModelTest 2: more models, new heuristics and parallel computing[J]. Nature Methods, 2012, 9(8): 772. [36] Rampal J. Biodiversité et biogéographie chez les Cavoliniidae (Mollusca, Gastropoda, Opisthobranchia, Euthecosomata). Régions faunistiques marine[J]. Zoosystema, 2002, 24(2): 209−258. [37] 陈钢. 海洋生物中的隐藏生物多样性与物种形成: 终生浮游生物悖论[J]. 厦门大学学报(自然科学版), 2006, 45(S2): 68−76.Chen Gang. Cryptic biodiversity and speciation in marine populations: the holoplankton paradox[J]. Journal of Xiamen University (Natural Science), 2006, 45(S2): 68−76. [38] Havermans C, Nagy Z T, Sonet G, et al. DNA barcoding reveals new insights into the diversity of Antarctic species of Orchomene sensu lato (Crustacea: Amphipoda: Lysianassoidea)[J]. Deep Sea Research II: Topical Studies in Oceanography, 2011, 58(1/2): 230−241. [39] Wall-Palmer D, Burridge A K, Goetze E, et al. Biogeography and genetic diversity of the atlantid heteropods[J]. Progress in Oceanography, 2018, 160: 1−25. doi: 10.1016/j.pocean.2017.11.004 [40] 刘泽. 鸟类线粒体假基因研究[D]. 大连: 辽宁师范大学, 2005.Liu Ze. Study of numt sequences among birds[D]. Dalian: Liaoning Normal University, 2005. -




 下载:
下载: