Screening of the reference genes of Skeletonema marinoi under different concentration of Fe3+ conditions in real-time quantitative PCR analysis
-
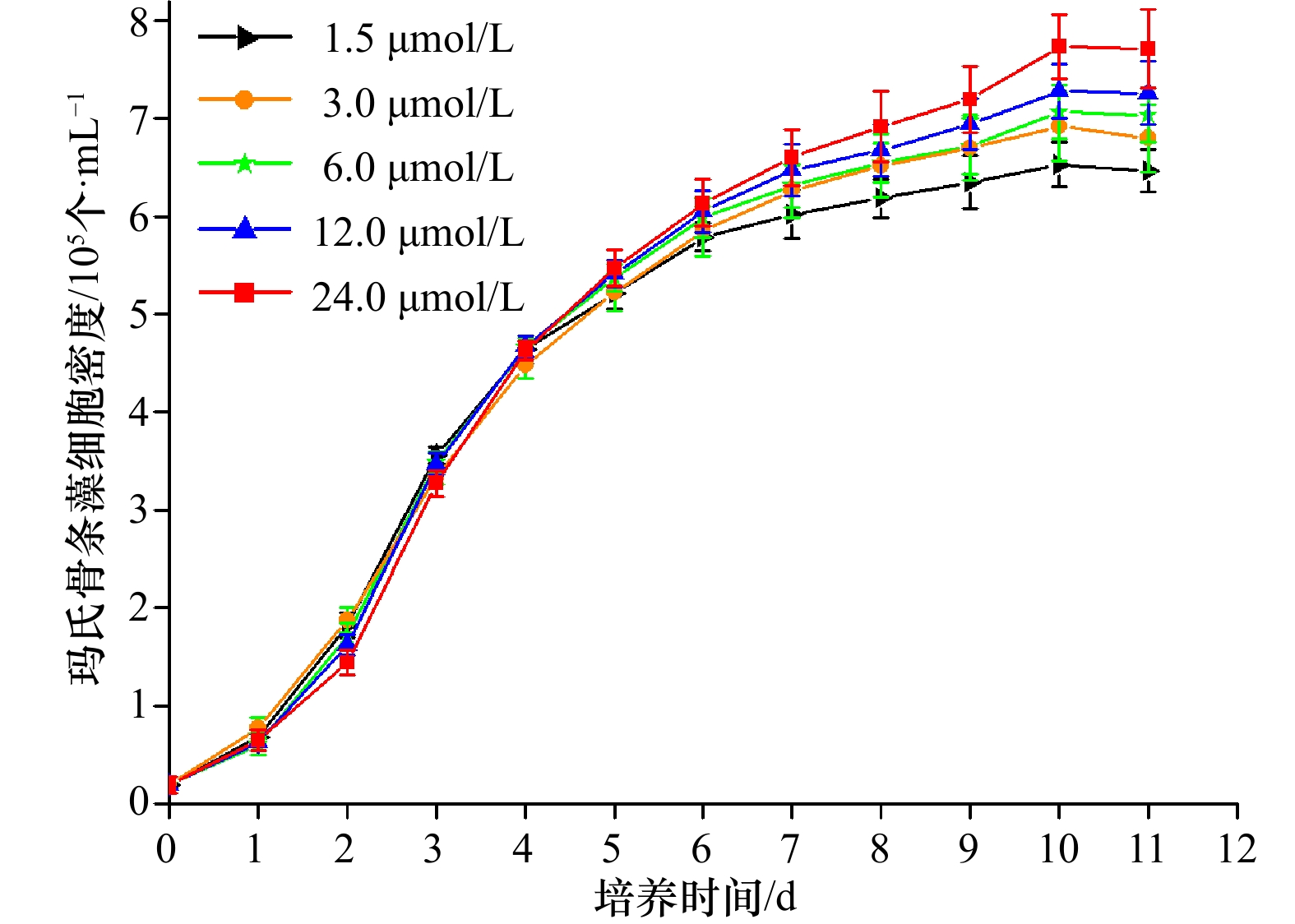
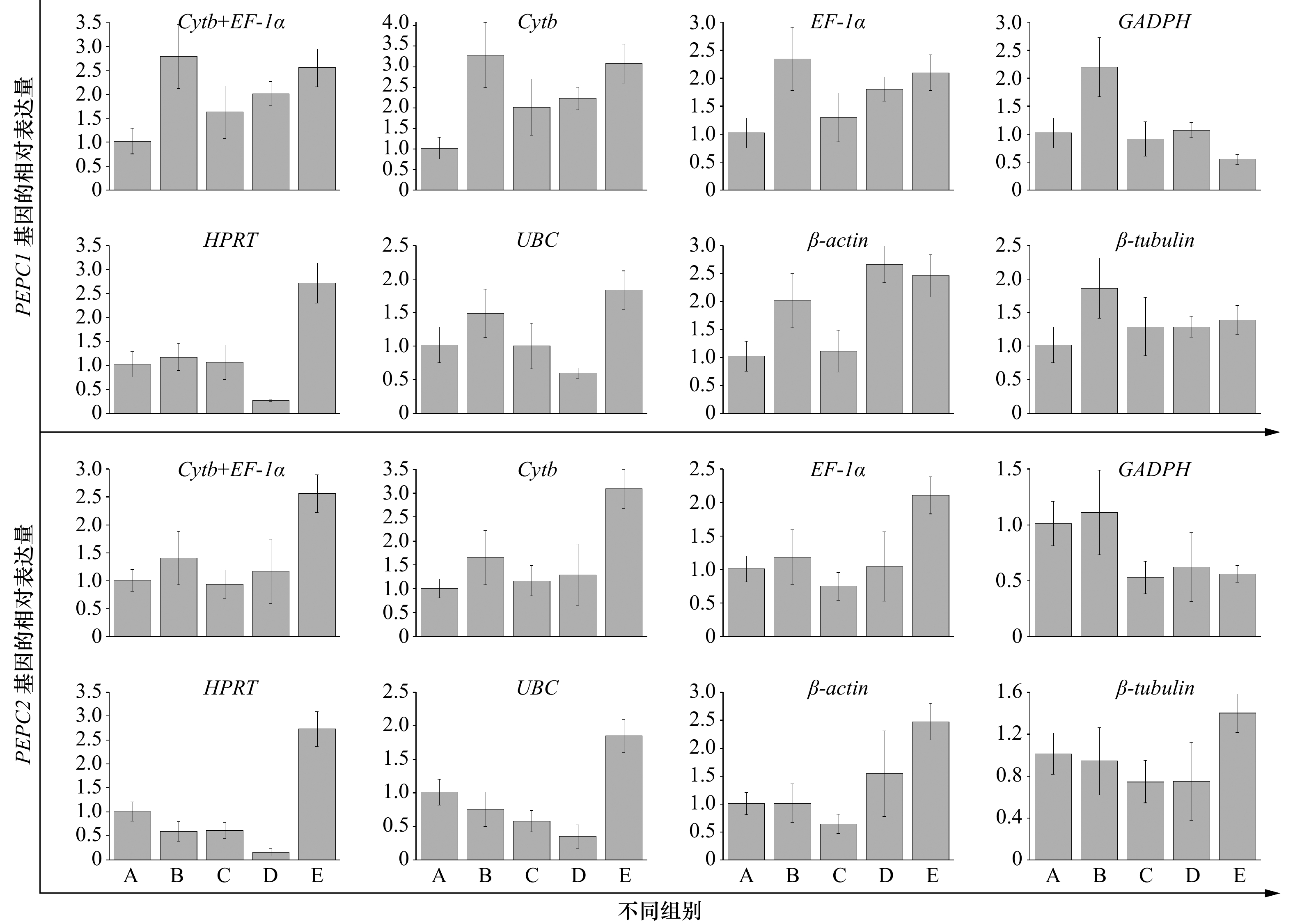
摘要: 实时荧光定量PCR(qRT-PCR)是定量分析基因表达的常用方法,选择合适的内参基因对准确分析目的基因表达水平至关重要。本研究以不同铁浓度培养条件下的玛氏骨条藻为材料,定量分析Cytb、EF-1α、HPRT、UBC、GAPDH、β-actin以及β-tubulin 7个内参基因的表达情况,并利用GeNorm、NormFinder和BestKeeper软件对这些内参基因的稳定性进行综合评价。结果表明,Cytb和EF-1α的表达稳定性较好,EF-1α+ Cytb组合的稳定性最佳,是玛氏骨条藻基因表达研究的理想内参基因,而其他基因的表达稳定性较差,不适合作为内参基因。本研究为玛氏骨条藻基因表达研究过程中内参基因的选择提供了方法学上的依据。Abstract: Real-time quantitative PCR (qRT-PCR) is a common method for quantitative analysis of gene expression. Selection of appropriate reference genes is essential for the accurate analysis of target gene expression levels. In this study, the expression of seven reference genes of Cytb, EF-1α, HPRT, UBC, GAPDH, β-actin and β-tubulin was quantitatively analyzed with different concentrations of iron concentration. The GeNorm, NormFinder and BestKeeper software comprehensively evaluated the stability of these reference genes. The results showed that the expression stability of Cytb and EF-1α was better, and the combination of EF-1α + Cytb was the best. It could be used as a reference gene for the study of gene expression in Skeletonema marinoi, while the expression stability of other genes was poor, and they were not suitable for being used as a reference gene. This study provides a methodological basis for the selection of reference genes during the study of gene expression in S. marinoi.
-
Key words:
- Fe3+ conditions /
- Skeletonema marinoi /
- reference genes /
- real-time quantitative PCR
-
表 1 qRT-PCR检测中9个基因的引物序列及其他相关信息
Tab. 1 Primer sequences and other relevant information of nine genes in qRT-PCR
基因名称 引物序列(5’-3’) AL/bp Tm/℃ AE/% R2 Cytb (F)GGCTTTGAGGGGGATTCACA
(R)AACGGGATTTTGTCACCAGT172 74.1 99.4 0.989 EF1-α (F)GAGCGTGAGCGTGGAGTTAC
(R)CGGCAGGCACAAGAAGAAG160 79.2 100.6 0.989 GAPDH (F)TCGGTATTAGGAACCCAGAGG
(R)TTGACGCCCACAACAAACAT178 77.9 99.0 0.988 HPRT (F)TTTGCGTCAGGGCTTTTACA
(R)CAGATTTGGGGTTGGCTTCT206 77 101.9 0.996 UBC (F)CGACCCAGCAAGTCCAAAG
(R)CCCATCGCTTCCCTCAAA152 78.8 101.2 0.984 β-actin (F)TCGTCGCCGTTGACTTTG
(R)ATTTCCTTGGACATACGCTCAC298 79.1 102.1 0.996 β-tubulin (F)ATCAACTCAAACGCATCAACG
(R)GTTATTACCCGCCCCACTCT176 81.3 99.2 0.998 PEPC1 (F)AGCGTGCTGGGCTCAATAT
(R)GCAGGATTACCTCCACGACC120 79.3 104.0 0.995 PEPC2 (F)GTTTCGGCATTTGGGCTTAC
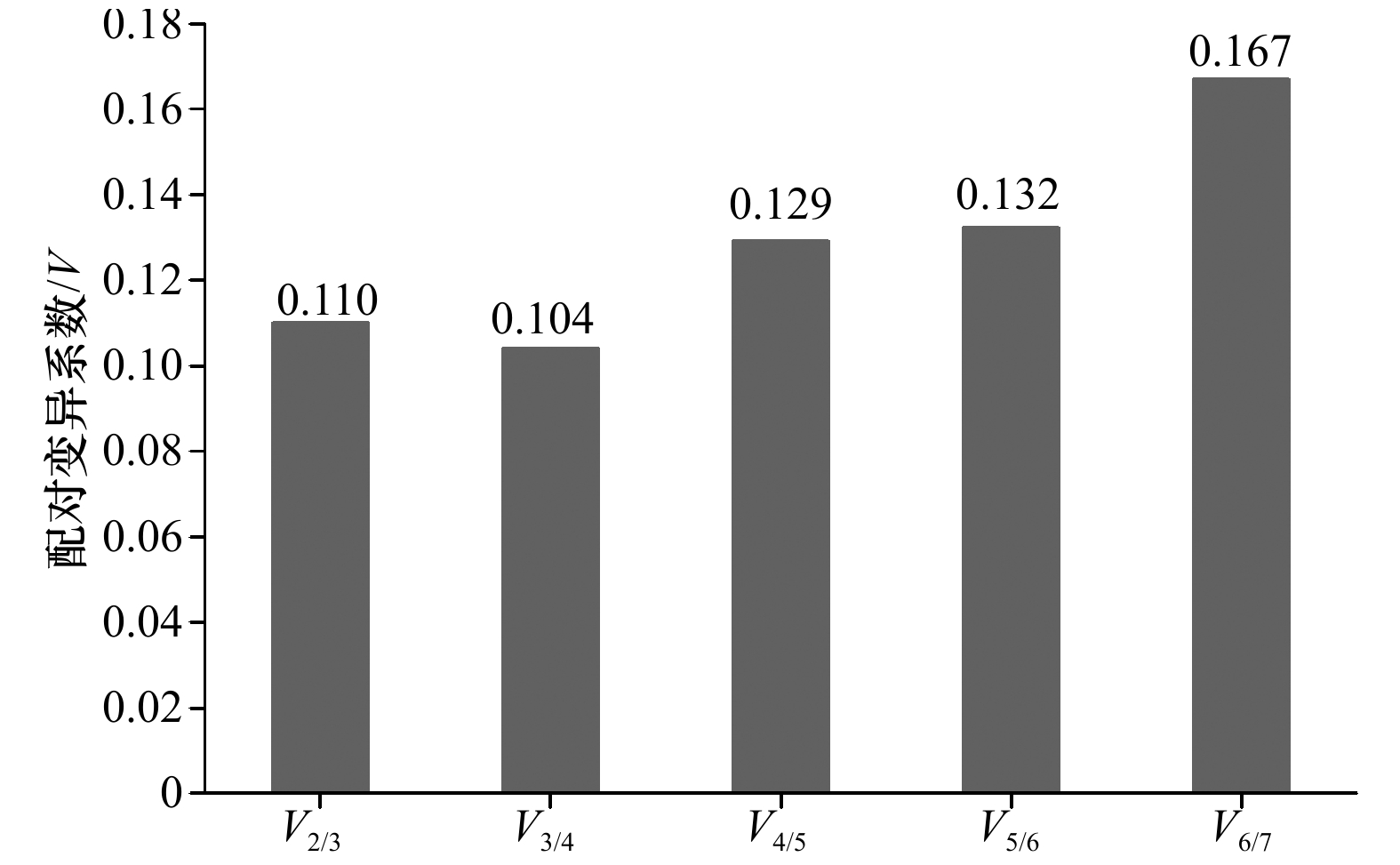
(R)ATTTCGCCATTGTCGTTCC209 78.9 101.0 0.997 注:AL为扩增长度;Tm为熔解温度;AE为引物的扩增效率;R2为确定系数。 表 2 NormFinder分析7个内参基因的表达稳定性指数
Tab. 2 The average expression stability value (M) of seven reference genes calculated by NormFinder
基因名称 M 稳定性排序 Cytb 0.240 3 EF1-α 0.084 1 GAPDH 0.585 6 HPRT 0.800 7 UBC 0.249 4 β-actin 0.409 5 β-tubulin 0.094 2 表 3 BestKeeper分析7个内参基因的表达稳定性指数
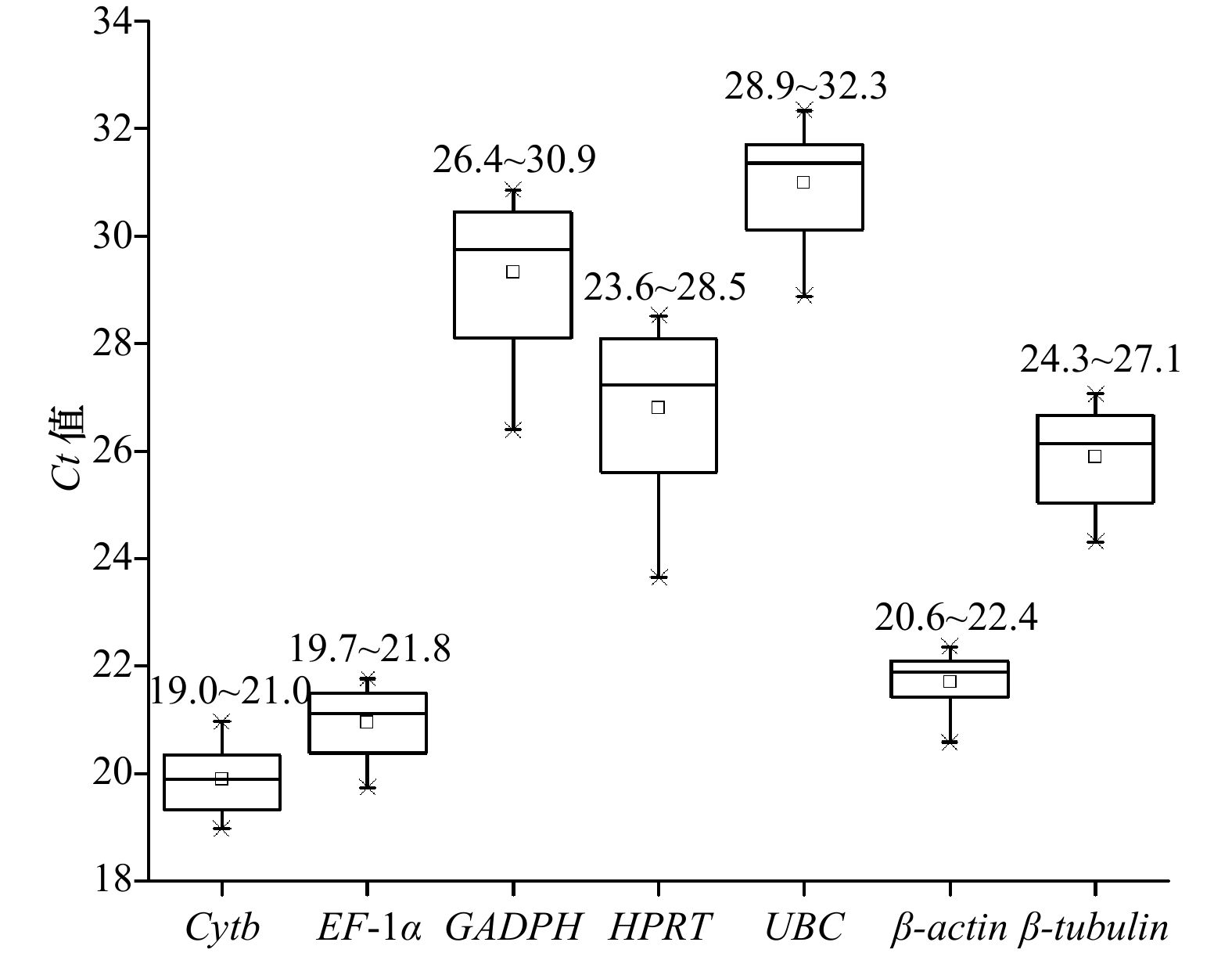
Tab. 3 The average expression stability value(M)of seven reference genes calculated by BestKeeper
基因名称 Cytb EF-1α GAPDH HPRT UBC β-actin β-tubulin 几何平均值 [Ct] 19.88 20.94 29.31 26.78 30.97 21.71 25.88 数算平均值 [Ct] 19.89 20.95 29.33 26.81 30.98 21.71 25.89 Min [Ct] 19.32 20.22 27.34 24.34 29.48 21.15 24.85 Max [Ct] 20.53 21.40 30.43 27.98 31.99 22.09 26.62 SD [± Ct] 0.46 0.48 1.03 0.99 0.74 0.23 0.69 CV [% Ct] 2.29 2.31 3.50 3.69 2.40 1.05 2.65 r 0.88 0.97 0.87 0.75 0.93 0.72 0.97 稳定性
排序2 3 7 6 5 1 4 表 4 7个内参基因的表达稳定性的统计分析
Tab. 4 Statistical analysis of the expression stability of seven reference genes
评价方法 Cytb EF-1α GAPDH HPRT UBC β-actin β-tubulin GeNorm 1 1 6 7 5 4 3 NormFinder 3 1 6 7 4 5 2 BestKeeper 2 3 7 6 5 1 4 Total 6 5 19 20 14 10 9 -
[1] 程金凤, 高亚辉, 梁君荣, 等. 骨条藻的种类与基因多样性研究进展[J]. 自然科学进展, 2007, 17(5): 586−594. doi: 10.3321/j.issn:1002-008X.2007.05.005Chen Jinfeng, Gao Yahui, Liang Junrong, et al. Advances in the research on the species and gene diversity of Skeletonema spp.[J]. Progress in Natural Science, 2007, 17(5): 586−594. doi: 10.3321/j.issn:1002-008X.2007.05.005 [2] 张晓东.厦门港骨条藻Skeletonema (Bacillariophyta) 物种多样性及周年变化的研究[D]. 青岛: 中国科学院海洋研究所, 2012.Diversity and annual variation of Skeletonema (Bacillariophyta) in Xiamen Harbor waters[D]. Qingdao: The Institute of Oceanology, Chinese Academy of Sciences, 2012. [3] Huggett J, Dheda K, Bustin S, et al. Real-time RT-PCR normalisation; strategies and considerations[J]. Genes & Immunity, 2005, 6(4): 279−284. [4] Udvardi M K, Czechowski T, Scheible W R. Eleven golden rules of quantitative RT-PCR[J]. The Plant Cell, 2008, 20(7): 1736−1737. doi: 10.1105/tpc.108.061143 [5] Vanguilder H D, Vrana K E, Freeman W M. Twenty-five years of quantitative PCR for gene expression analysis[J]. Biotechniques, 2008, 44(5): 619−626. doi: 10.2144/000112776 [6] Quackenbush J. Microarray data normalization and transformation[J]. Nature Genetics, 2002, 32(S1): 496−501. [7] Nolan T, Hands R E, Bustin S A. Quantification of mRNA using real-time RT-PCR[J]. Nature Protocols, 2006, 1(3): 1559−1582. doi: 10.1038/nprot.2006.236 [8] Albershardt T C, Iritani B M, Ruddell A. Evaluation of reference genes for quantitative PCR analysis of mouse lymphocytes[J]. Journal of Immunological Methods, 2012, 384(1/2): 196−199. [9] Mohelnikova-Duchonova B, Oliverius M, Honsova E, et al. Evaluation of reference genes and normalization strategy for quantitative real-time PCR in human pancreatic carcinoma[J]. Disease Markers, 2012, 32(3): 203−210. doi: 10.1155/2012/582107 [10] Luo Huolin, Luo Kecan, Luo Liping, et al. Evaluation of candidate reference genes for gene expression studies in Cymbidium kanran[J]. Scientia Horticulturae, 2014, 167: 43−48. doi: 10.1016/j.scienta.2013.12.030 [11] Adelfi M G, Borra M, Sanges R, et al. Selection and validation of reference genes for qPCR analysis in the pennate diatoms Pseudo-nitzschia multistriata and P. arenysensis[J]. Journal of Experimental Marine Biology and Ecology, 2014, 451: 74−81. doi: 10.1016/j.jembe.2013.11.003 [12] Cao Shaona, Zhang Xiaowen, Ye Naihao, et al. Evaluation of putative internal reference genes for gene expression normalization in Nannochloropsis sp. by quantitative real-time RT-PCR[J]. Biochemical and Biophysical Research Communications, 2012, 424(1): 118−123. doi: 10.1016/j.bbrc.2012.06.086 [13] Dong Meitao, Zhang Xiaowen, Chi Xiaoyuan, et al. The validity of a reference gene is highly dependent on the experimental conditions in green alga Ulva linza[J]. Current Genetics, 2012, 58(1): 13−20. doi: 10.1007/s00294-011-0361-3 [14] Briat J F, Curie C, Gaymard F. Iron utilization and metabolism in plants[J]. Current Opinion in Plant Biology, 2007, 10(3): 276−282. doi: 10.1016/j.pbi.2007.04.003 [15] Greene R M, Geider R J, Kolber Z, et al. Iron-induced changes in light harvesting and photochemical energy conversion processes in eukaryotic marine algae[J]. Plant Physiology, 1992, 100(2): 565−575. doi: 10.1104/pp.100.2.565 [16] Vassiliev I R, Kolber Z, Wyman K D, et al. Effects of iron limitation on photosystem Ⅱ. composition and light utilization in Dunaliella tertiolecta[J]. Plant Physiology, 1995, 109(3): 963−972. doi: 10.1104/pp.109.3.963 [17] Ivanov A, Park Y I, Miskiewicz E, et al. Iron stress restricts photosynthetic intersystem electron transport in Synechococcus sp. PCC 7942[J]. FEBS Letters, 2000, 485(2/3): 173−177. [18] Wan Minxi, Jin Xuejie, Xia Jinlan, et al. The effect of iron on growth, lipid accumulation, and gene expression profile of the freshwater microalga Chlorella sorokiniana[J]. Applied Microbiology and Biotechnology, 2014, 98(22): 9473−9481. doi: 10.1007/s00253-014-6088-6 [19] Whitney L P, Lins J J, Hughes M P, et al. Characterization of putative iron responsive genes as species-specific indicators of iron stress in Thalassiosiroid diatoms[J]. Frontiers in Microbiology, 2011, 2: 234. [20] Hernández-Torres A, Zapata-Morales A L, Alfaro A E O, et al. Identification of gene transcripts involved in lipid biosynthesis in Chlamydomonas reinhardtii under nitrogen, iron and sulfur deprivation[J]. World Journal of Microbiology and Biotechnology, 2016, 32(4): 55. doi: 10.1007/s11274-016-2008-5 [21] Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biology, 2002, 3(7): research0034.1. [22] Andersen C L, Jensen J L, Ørntoft T F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets[J]. Cancer Research, 2004, 64(15): 5245−5250. doi: 10.1158/0008-5472.CAN-04-0496 [23] Pfaffl M W, Tichopad A, Prgomet C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper–excel-based tool using pair-wise correlations[J]. Biotechnology Letters, 2004, 26(6): 509−515. doi: 10.1023/B:BILE.0000019559.84305.47 [24] Livak K J, Schmittgen T D. Analysis of relative gene expression data using real-time quantitative PCR and the method[J]. Methods, 2001, 25(4): 402−408. doi: 10.1006/meth.2001.1262 [25] Wan Hongjian, Zhao Zhenguo, Qian Chuntao, et al. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber[J]. Analytical Biochemistry, 2010, 399(2): 257−261. doi: 10.1016/j.ab.2009.12.008 [26] Huis R, Hawkins S, Neutelings G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.)[J]. BMC Plant Biology, 2010, 10: 71. doi: 10.1186/1471-2229-10-71 [27] Mafra V, Kubo K S, Alves-Ferreira M, et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions[J]. PLoS One, 2012, 7(2): e31263. doi: 10.1371/journal.pone.0031263 [28] Dheda K, Huggett J F, Chang J S, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization[J]. Analytical Biochemistry, 2005, 344(1): 141−143. doi: 10.1016/j.ab.2005.05.022 [29] Zhu Jiang, He Fuhong, Song Shuhui, et al. How many human genes can be defined as housekeeping with current expression data?[J]. BMC Genomics, 2008, 9: 172. doi: 10.1186/1471-2164-9-172 [30] Schmid H, Cohen C D, Henger A, et al. Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies[J]. Kidney International, 2003, 64(1): 356−360. doi: 10.1046/j.1523-1755.2003.00074.x [31] 盖民昊, 陈堑, 胡祖庆, 等. 紫外诱导下麦长管蚜细胞色素b基因和SOD基因的克隆与序列分析[J]. 西北农林科技大学学报: 自然科学版, 2010, 38(11): 167−172.Gai Minhao, Chen Qian, Hu Zuqing, et al. Cloning and sequence analysis of cytochrome b gene and SOD gene in Sitobium avenae under the ultraviolet induction[J]. Journal of Northwest A&F University: Natural Science Edition, 2010, 38(11): 167−172. [32] 何闪英, 于志刚, 米铁柱. 增殖细胞核抗原基因表达量与中肋骨条藻生长的关系[J]. 水生生物学报, 2009, 33(1): 103−112.He Shanying, Yu Zhigang, Mi Tiezhu. Relationship between proliferating cell nuclear antigen gene expression amount and growth rate of Skeletonema costatum[J]. Acta Hydrobiologica Sinica, 2009, 33(1): 103−112. [33] 周冰, 曹诚, 刘传喧. 翻译延伸因子1A的研究进展[J]. 生物技术通讯, 2007, 18(2): 281−284. doi: 10.3969/j.issn.1009-0002.2007.02.032Zhou Bing, Cao Cheng, Liu Chuanxuan. Advances in research on translation elongation factor 1 alpha[J]. Letters in Biotechnology, 2007, 18(2): 281−284. doi: 10.3969/j.issn.1009-0002.2007.02.032 [34] Han Xiaojiao, Lu Mengzhu, Chen Yicun, et al. Selection of reliable reference genes for gene expression studies using real-time PCR in tung tree during seed development[J]. PLoS One, 2012, 7(8): e43084. doi: 10.1371/journal.pone.0043084 [35] Martin R C, Hollenbeck V G, Dombrowski J E. Evaluation of reference genes for quantitative RT-PCR in Lolium perenne[J]. Crop Science, 2008, 48(5): 1881−1887. doi: 10.2135/cropsci2007.10.0597 [36] Basa B, Solti Á, Sárvári É, et al. Housekeeping gene selection in poplar plants under Cd-stress: comparative study for real-time PCR normalisation[J]. Functional Plant Biology, 2009, 36(12): 1079−1087. doi: 10.1071/FP09073 [37] 蔡文凯, 胡金璐, 李双双, 等. 辐射条件下微藻基因表达内参基因的选择[J]. 空间科学学报, 2013, 33(6): 651−658. doi: 10.11728/cjss2013.06.651Cai Wenkai, Hu Jinlu, Li Shuangshuang, et al. Selection of suitable internal control genes in microalgae under radiation condition[J]. Chinese Journal of Space Science, 2013, 33(6): 651−658. doi: 10.11728/cjss2013.06.651 [38] Hong S Y, Seo P J, Yang M S, et al. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR[J]. BMC Plant Biology, 2008, 8: 112. doi: 10.1186/1471-2229-8-112 [39] 苏晓娟, 樊保国, 袁丽钗, 等. 实时荧光定量PCR分析中毛果杨内参基因的筛选和验证[J]. 植物学报, 2013, 48(5): 507−518.Su Xiaojuan, Fan Baoguo, Yuan Lichai, et al. Selection and validation of reference genes for quantitative RT-PCR analysis of gene expression in Populus trichocarpa[J]. Bulletin of Botany, 2013, 48(5): 507−518. [40] 吴文凯, 刘成前, 周志刚, 等. 用于莱茵衣藻荧光定量PCR分析的内参基因选择[J]. 植物生理学通讯, 2009, 45(7): 667−672.Wu Wenkai, Liu Chengqian, Zhou Zhigang, et al. The selection of reference genes in Chlamydomonas reinhardtii P. A. dangeard by real-time quantitative PCR[J]. Plant Physiology Communications, 2009, 45(7): 667−672. [41] Shim J, Shim E, Kim G H, et al. Keeping house: evaluation of housekeeping genes for real-time PCR in the red alga, Bostrychia moritziana (Florideophyceae)[J]. Algae, 2016, 31(2): 167−174. doi: 10.4490/algae.2016.31.5.25 [42] Liu Chenlin, Wu Guangting, Huang Xiaohang, et al. Validation of housekeeping genes for gene expression studies in an ice alga Chlamydomonas during freezing acclimation[J]. Extremophiles, 2012, 16(3): 419−425. doi: 10.1007/s00792-012-0441-4 [43] Wu Shuang, Zhou Jiannan, Cao Xupeng, et al. Determination of internal controls for quantitative gene expression of Isochrysis zhangjiangensis at nitrogen stress condition[J]. Journal of Ocean University of China, 2016, 15(1): 137−144. doi: 10.1007/s11802-016-2847-6 -





 下载:
下载: