Taxonomy and phylogeography of Clio species based on mtCOI and 18S rRNA genes
-
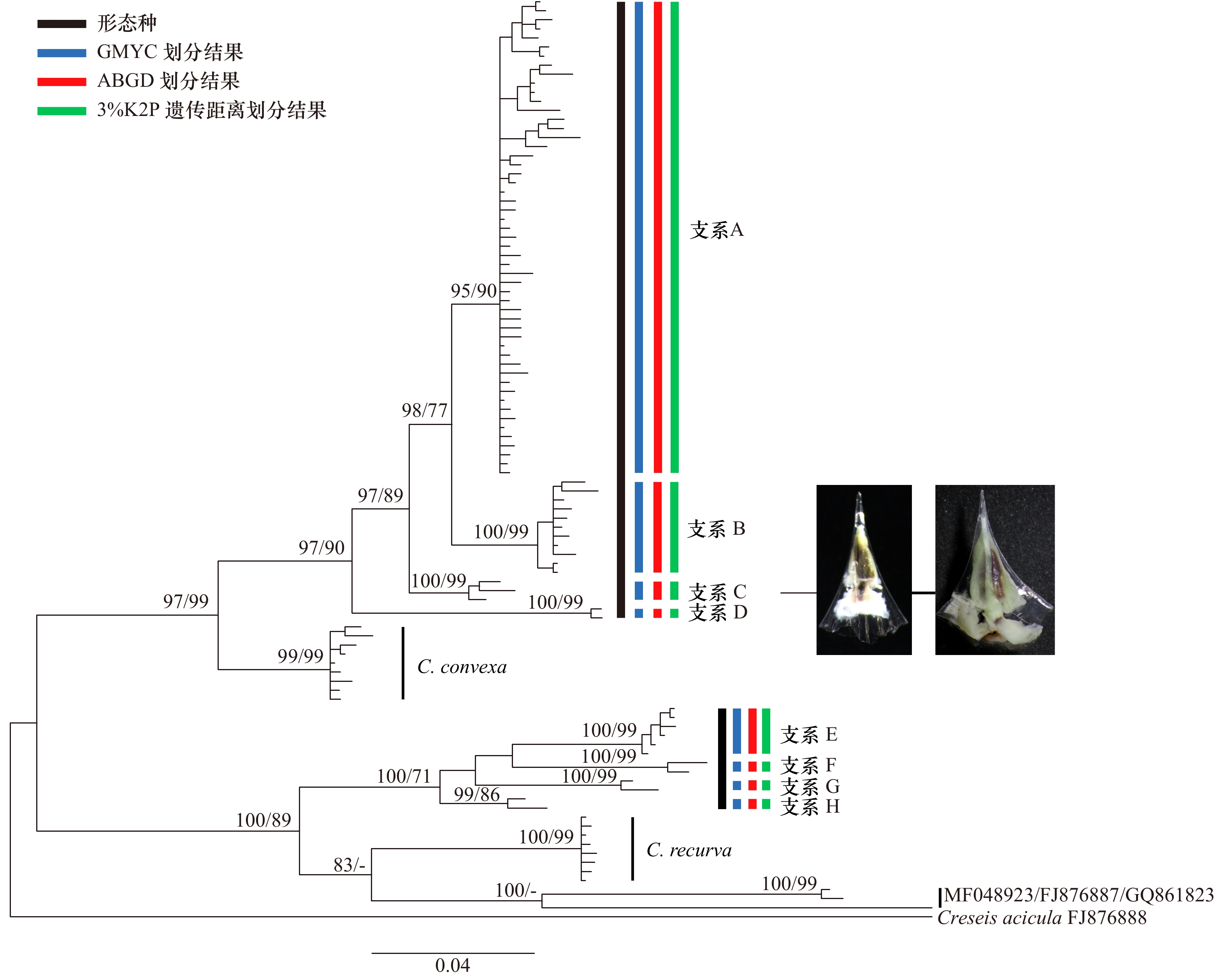
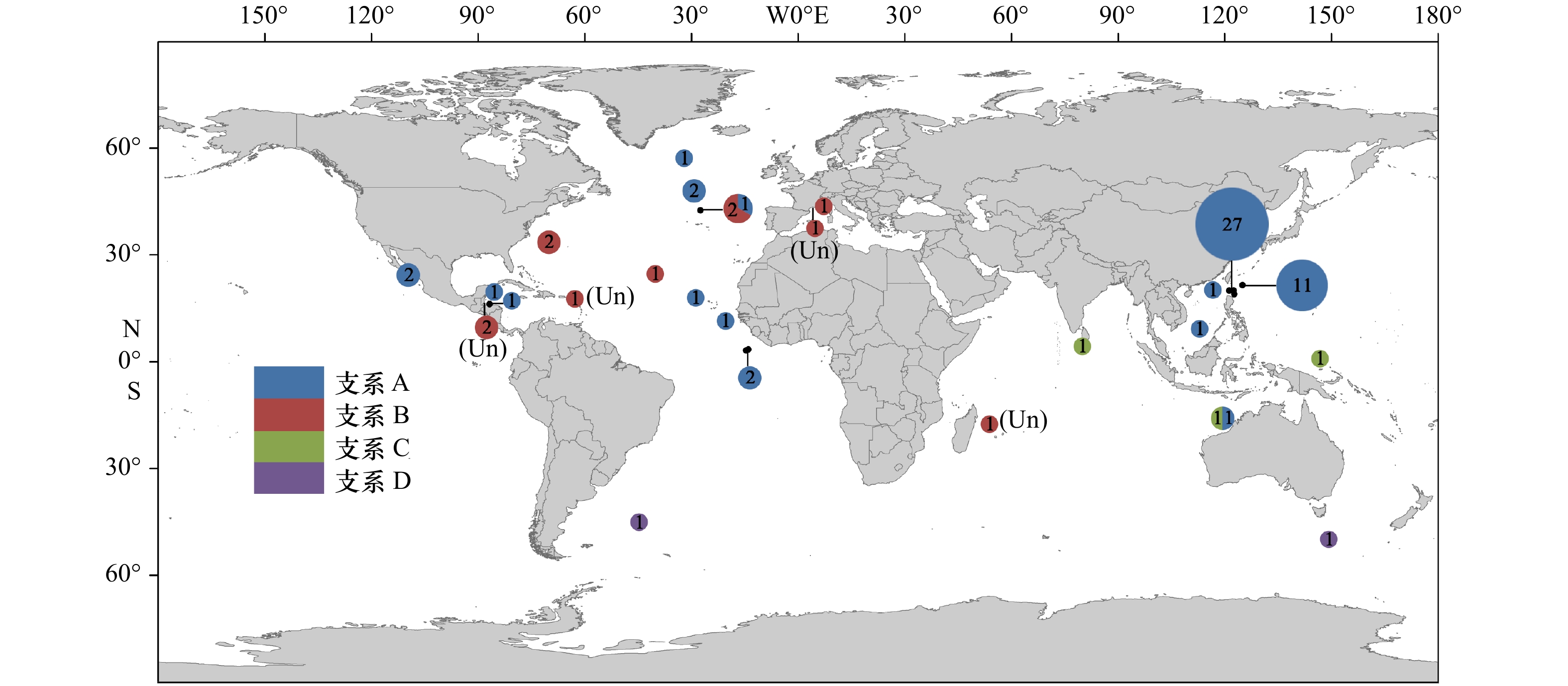
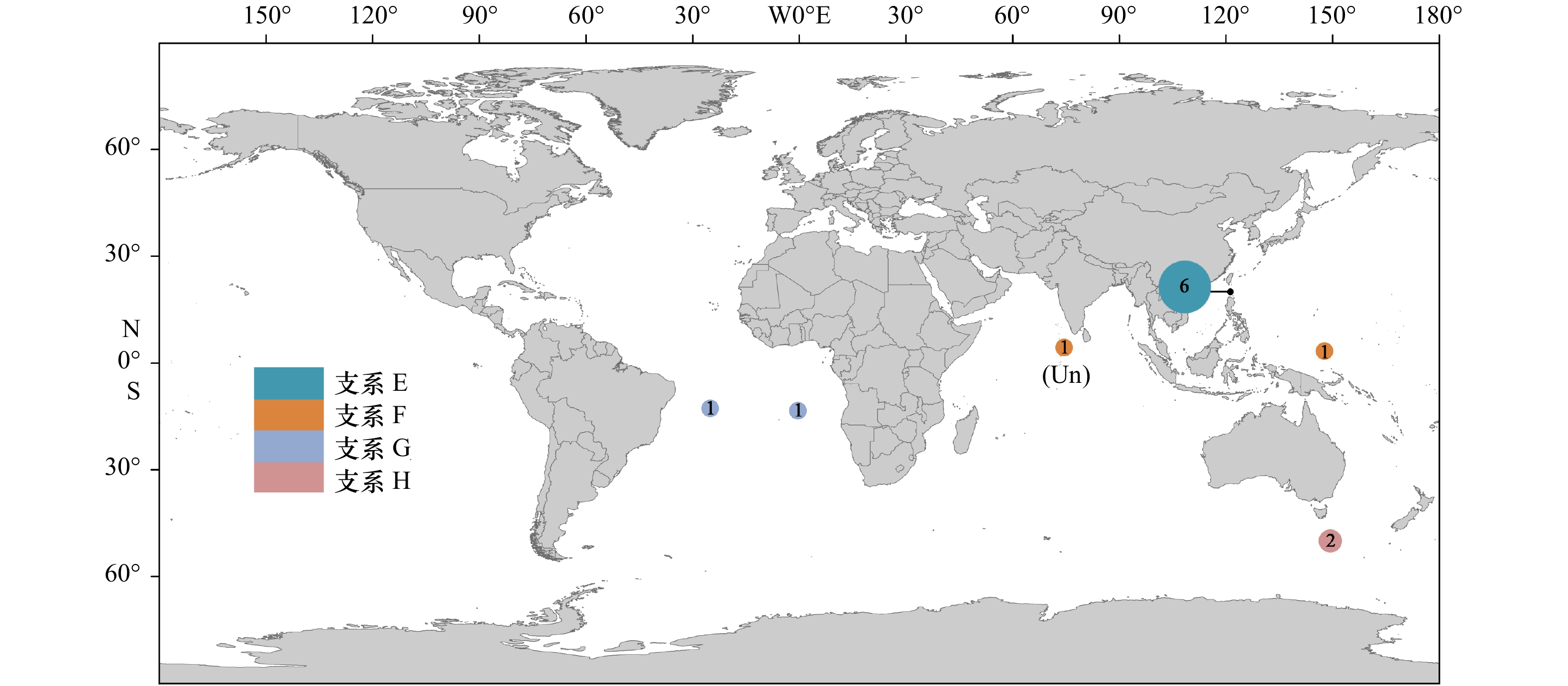
摘要: 许多终生浮游软体动物接近全球分布或呈环球分布,是海洋酸化和谱系地理学研究的良好材料。本文以西北太平洋和北印度洋的长角螺属(Clio)种类为材料,通过测定其线粒体COI基因(mtCOI,55条)和核18S rRNA基因(9条)序列,结合数据库中已有的序列,对该属进行了分类学和谱系地理学研究。结果表明,矛头长角螺(C. pyramidata)和尖棘长角螺(C. cuspidata)在mtCOI基因系统树中均形成4个明显分化的谱系分支,分别为支系A–D和支系E–H。矛头长角螺的支系A为全球分布,中国海及邻近海域可能仅有支系A的存在,支系B、C和D分布于特定海域。尖棘长角螺也存在明显的谱系地理结构,西北太平洋北赤道流南北两侧存在2个不同的支系,分布于吕宋海峡的支系E为一新的谱系分支。各支系内mtCOI基因的K2P遗传距离在0~0.026之间,支系间的遗传距离在0.031~0.089之间。膨凸长角螺(C. convexa)和曲形长角螺(C. recurva)没有明显的地理遗传分化。18S rRNA基因支持支系D为独立种,但不支持其他支系的划分。矛头长角螺和尖棘长角螺内部可能存在隐存多样性。洋流可能会成为物种扩布和基因交流的障碍。Abstract: Many species of holoplanktonic gastropods have a near-cosmopolitan or circumglobal distribution, some of which are used for phylogeography and ocean acidification research. Using mitochondrial COI (mtCOI, 55 animals) and nuclear 18S rRNA (9 animals) sequence data from specimens sampled from Northwest Pacific and North Indian Oceans, and together with the sequences from GenBank, we investigated the taxonomy and phylogeography of Clio species. Four lineages were defined by mtCOI tree for C. pyramidata (Lineages A–D) and C. cuspidata (LineagesE–H), respectively. Lineage A of C. pyramidata showed a circumglobal distribution, while Lineages B, C and D had their restrained dispersal. Only Lineage A was found in Chinese seas and adjacent waters. The divergent mtCOI lineages of C. cuspidata were also phylogeographical structured. Two distinct lineages (E and F) of C. cuspidata were observed in the Northwestern Pacific, which are distributed in the south and north areas of the North Equatorial Current respectively. The K2P distances of intra-lineage were between 0 and 0.026, while the distances of inter-lineage ranged from 0.031 to 0.089. Geographic structures were not observed in C. convexa and C. recurva. The morphospecies C. pyramidata and C. cuspidata may harbour cryptic diversity. Our results also suggested that ocean currents may limit the species dispersal and gene flow.
-
Key words:
- Clio /
- taxonomy /
- phyogeography /
- mtCOI /
- 18S rRNA
-
图 1 长角螺属种类的贝壳形态(比例尺=5 mm)
a-c. 矛头长角螺(支系A); d-e. 尖棘长角螺(支系E); f. 尖棘长角螺(支系F); g. 膨凸长角螺; h. 矛头长角螺(幼体, 支系C)
Fig. 1 Shell morphology of Clio species (scale bars=5 mm)
a-c. C. pyramidata, Lineage A); d-e. C. cuspidate (Lineage E); f. C. cuspidata (Lineage F); g. C. convexa; h. C. pyramidata (juvenile, Lineage C)
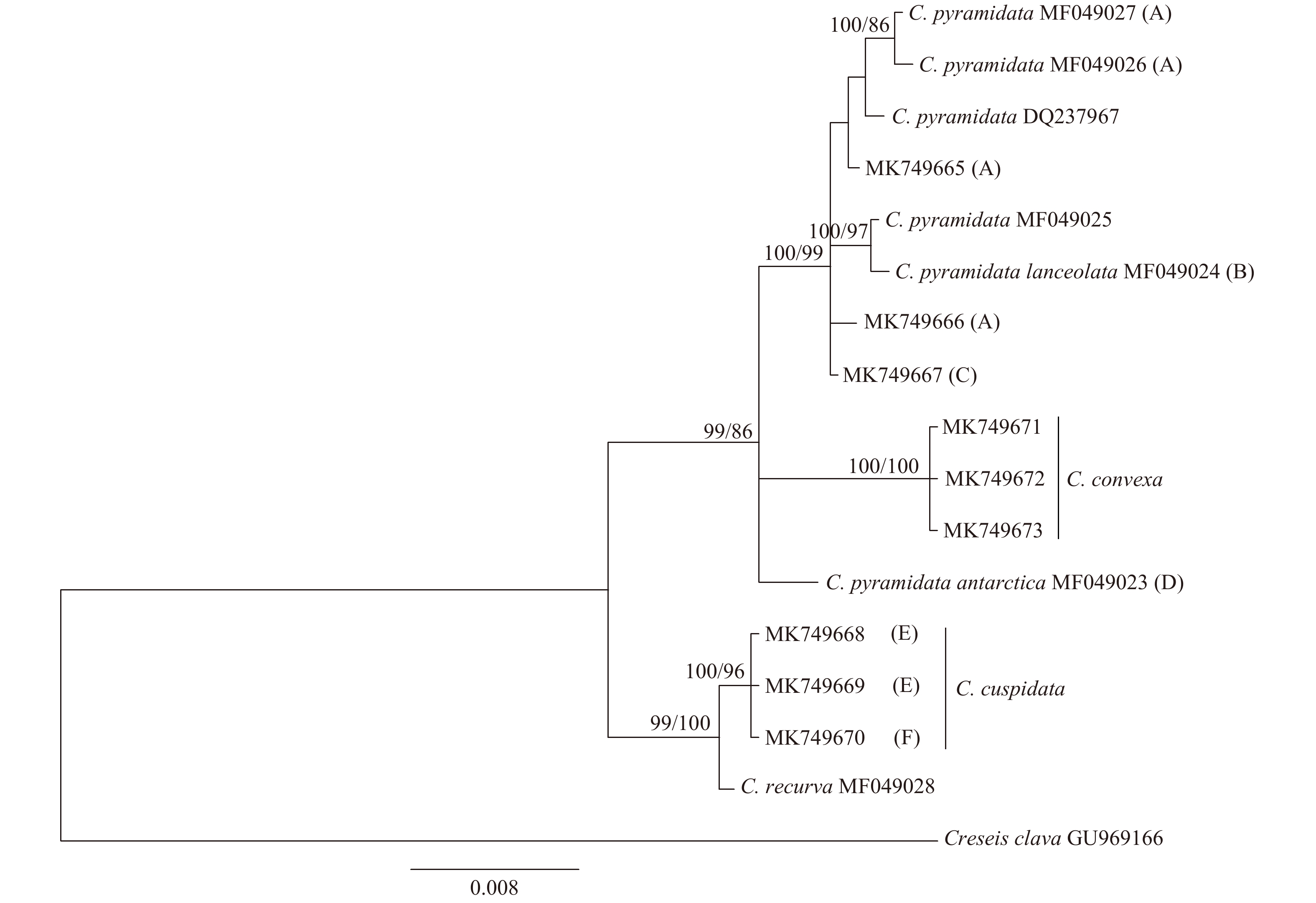
图 3 基于18S rRNA基因构建的长角螺属贝叶斯系统发育树
节点处数值分别为后验概率和自展支持率, 仅给出大于70%的数值; 括号中的大写字母对应于mtCOI的支系编号
Fig. 3 Bayesian inference phylogenetic tree of genus Clio based on nuclear18S rRNA gene sequences
Posterior probabilities/bootstrap values over 70% are shown for each node; capital letters in brackets are corresponding to mtCOI lineages
表 1 测序标本的采样信息
Tab. 1 Collection information for specimens analyzed in this study
种类 标本编号 标本数量 经纬度 采样时间 GenBank登录号 mtCOI 18S rRNA C. pyramidata MT1 1 19.000°N, 122.667°E 2016−06−01 MK749610 C. pyramidata MT2 1 20.000°N, 122.500°E 2017−09−11 MK749611 MK749665 C. pyramidata MT3~MT27 25 20.000°N, 121.250°E 2018−05−07 MK749612~36 MK749666 C. pyramidata MT28~MT38 11 21.500°N, 125.000°E 2016−05−29 MK749637~47 C. pyramidata MT39 1 20.260°N, 116.576°E 2016−06−04 MK749648 C. pyramidata MT40 1 9.470°N, 112.870°E 2017−07−02 MK749649 C. pyramidata MT41 1 0.929°N, 146.824°E 2018−12−07 MK749650 C. pyramidata MT42 1 4.507°N, 80.044°E 2018−04−04 MK749651 MK749667 C. cuspidata JJ1~JJ6 6 20.000°N, 121.250°E 2016−06−02/2018−05−07 MK749652~57 MK749668~69 C. cuspidata JJ7 1 3.210°N, 147.945°E 2019−01−02 MK749658 MK749670 C. convexa PT1 1 10.000°N, 87.501°E 2018−04−29 MK749659 MK749671 C. convexa PT2 1 0.934°N, 149.658°E 2018−12−13 MK749660 MK749672 C. convexa PT3~PT6 4 3.768°N, 146.820°E 2019−01−01 MK749661~64 MK749673 表 2 基于mtCOI基因的长角螺属4个形态种的K2P遗传距离
Tab. 2 The K2P distances of four Clio morphospecies based on mtCOI sequences
形态种 C. pyramidata C. convexa C. recurva C. cuspidata C. pyramidata 0~0.089(0.025) C. convexa 0.070~0.113(0.089) 0.002~0.019(0.008) C. recurva 0.153~0.185(0.170) 0.131~0.148(0.139) 0~0.005(0.002) C. cuspidata 0.133~0.183(0.156) 0.125~0.168(0.141) 0.083~0.110(0.097) 0~0.079(0.049) 注:括号中的数值为平均值 -
[1] Bednaršek N, Možina J, Vučković M, et al. Global distribution of pteropods representing carbonate functional type biomass[J]. Earth System Science Data Discussions, 2012, 5(1): 317−350. doi: 10.5194/essdd-5-317-2012 [2] Hunt B P V, Pakhomov E A, Hosie G W, et al. Pteropods in southern ocean ecosystems[J]. Progress in Oceanography, 2008, 78(3): 193−221. doi: 10.1016/j.pocean.2008.06.001 [3] Flores H, van Franeker J A, Cisewski B, et al. Macrofauna under sea ice and in the open surface layer of the Lazarev Sea, Southern Ocean[J]. Deep Sea Research Part II: Topical Studies in Oceanography, 2011, 58(19/20): 1948−1961. [4] Howard W R, Roberts D, Moy A D, et al. Distribution, abundance and seasonal flux of pteropods in the Sub-Antarctic zone[J]. Deep Sea Research Part II: Topical Studies in Oceanography, 2011, 58(21/22): 2293−2300. [5] Bednaršek N, Harvey C J, Kaplan I C, et al. Pteropods on the edge: cumulative effects of ocean acidification, warming, and deoxygenation[J]. Progress in Oceanography, 2016, 145: 1−24. doi: 10.1016/j.pocean.2016.04.002 [6] Fabry V J, Seibel B A, Feely R A, et al. Impacts of ocean acidification on marine fauna and ecosystem processes[J]. ICES Journal of Marine Science, 2008, 65(3): 414−432. doi: 10.1093/icesjms/fsn048 [7] Gardner J, Manno C, Bakker D C E, et al. Southern Ocean pteropods at risk from ocean warming and acidification[J]. Marine Biology, 2018, 165(1): 8. doi: 10.1007/s00227-017-3261-3 [8] 张福绥. 中国近海的浮游软体动物. I. 翼足类、异足类及海蜗牛类的分类研究[J]. 海洋科学集刊, 1964(5): 125−226.Zhang Fusui. The pelagic molluscs off China coast Ⅰ. A systemic study of Pteropoda (Opisthobranchia), Heteropoda (Prosobranchia) and Janthinidae (Ptenoglossa, Prosobranchia)[J]. Studia Marina Sinica, 1964(5): 125−226. [9] Robertson R. Dispersal and wastage of larval Philippia krebsii (Gastropoda: Architectonicidae) in the north Atlantic[J]. Proceedings of the Academy of Natural Sciences of Philadelphia, 1964, 116: 1−27. [10] Bieler R. Architectonicidae of the Indo-Pacific (Mollusca, Gastropoda)[M].Stuttgart: Abhandlungen des Naturwissenschaftlichen Vereins in Hamburg, 1993. [11] Bontes B, van der Spoel S. Variation in Diacria trispinosa group, new interpretation of colour patterns and description of D. rubecula n. sp. (Pteropoda)[J]. Bulletin Zoölogisch Museum, Universiteit van Amsterdam, 1998, 16(11): 77−84. [12] Hebert P D N, Cywinska A, Ball S L, et al. Biological identifications through DNA Barcodes[J]. Proceedings of the Royal Society B: Biological Sciences, 2003, 270(1512): 313−321. doi: 10.1098/rspb.2002.2218 [13] Jennings R M, Bucklin A, Ossenbrügger H, et al. Species diversity of planktonic gastropods (Pteropoda and Heteropoda) from six ocean regions based on DNA barcode analysis[J]. Deep Sea Research Part Ⅱ: Topical Studies in Oceanography, 2010, 57(24/26): 2199−2210. [14] Maas A E, Blanco-Bercial L, Lawson G L. Reexamination of the species assignment of Diacavolinia pteropods using DNA barcoding[J]. PLoS One, 2013, 8(1): e53889. doi: 10.1371/journal.pone.0053889 [15] Gasca R, Janssen A W. Taxonomic review, molecular data and key to the species of Creseidae from the Atlantic Ocean[J]. Journal of Molluscan Studies, 2014, 80(1): 35−42. doi: 10.1093/mollus/eyt038 [16] Wall-Palmer D, Burridge A K, Goetze E, et al. Biogeography and genetic diversity of the atlantid heteropods[J]. Progress in Oceanography, 2018, 160: 1−25. doi: 10.1016/j.pocean.2017.11.004 [17] Burridge A K, Goetze E, Raes N, et al. Global biogeography and evolution of Cuvierina pteropods[J]. BMC Evolutionary Biology, 2015, 15: 39. doi: 10.1186/s12862-015-0310-8 [18] 李海涛, 何薇, 周鹏, 等. 伶鼬榧螺(Oliva mustelina)的分子鉴定及其形态变异[J]. 海洋学报, 2015, 37(4): 117−123.Li Haitao, He Wei, Zhou Peng, et al. Molecular identification of Oliva mustelina and its morphological variation[J]. Haiyang Xuebao, 2015, 37(4): 117−123. [19] Folmer O, Black M, Heah W, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates[J]. Molecular Marine Biology and Biotechnology, 1994, 3(5): 294−299. [20] Vonnemann V, Schrödl M, Klussmann-Kolb A, et al. Reconstruction of the phylogeny of the Opisthobranchia (Mollusca: Gastropoda) by means of 18s and 28s rRNA gene sequences[J]. Journal of Molluscan Studies, 2005, 71(2): 113−125. doi: 10.1093/mollus/eyi014 [21] Ronquist F, Teslenko M, van der Mark P, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space[J]. Systematic Biology, 2012, 61(3): 539−452. doi: 10.1093/sysbio/sys029 [22] Darriba D, Taboada G L, Doallo R, et al. jModelTest 2: more models, new heuristics and parallel computing[J]. Nature Methods, 2012, 9(8): 772. [23] Fujisawa T, Barraclough T G. Delimiting species using single-locus data and the generalized Mixed Yule coalescent approach: a revised method and evaluation on simulated data sets[J]. Systematic Biology, 2013, 62(5): 707−724. doi: 10.1093/sysbio/syt033 [24] Puillandre N, Lambert A, Brouillet S, et al. ABGD, automatic barcode gap discovery for primary species delimitation[J]. Molecular Ecology, 2012, 21(8): 1864−1877. doi: 10.1111/j.1365-294X.2011.05239.x [25] Drummond A J, Rambaut A. BEAST: bayesian evolutionary analysis by sampling trees[J]. BMC Evolutionary Biology, 2007, 7: 214. doi: 10.1186/1471-2148-7-214 [26] van der Spoel S, Dadon J R. Pteropoda[M]//Boltovskoy D. South Atlantic zooplankton. Leiden, The Netherlands: Backhuys, 1999: 649−706. [27] Palumbi S R. Genetic divergence, reproductive isolation, and marine speciation[J]. Annual Review of Ecology and Systematics, 1994, 25: 547−572. doi: 10.1146/annurev.es.25.110194.002555 [28] Chen Gang, Hare M P. Cryptic diversity and comparative phylogeography of the estuarine copepod Acartia tonsa on the US Atlantic coast[J]. Molecular Ecology, 2011, 20(11): 2425−2441. doi: 10.1111/j.1365-294X.2011.05079.x [29] Peijnenburg K T C A, Goetze E. High evolutionary potential of marine zooplankton[J]. Ecology and Evolution, 2013, 3(8): 2765−2781. doi: 10.1002/ece3.644 [30] Goetze E, Hüdepohl P T, Chang C, et al. Ecological dispersal barrier across the equatorial Atlantic in a migratory planktonic copepod[J]. Progress in Oceanography, 2017, 158: 203−212. doi: 10.1016/j.pocean.2016.07.001 [31] Toole J M, Millard R C, Wang Z, et al. Observations of the pacific north equatorial current bifurcation at the Philippine coast[J]. Journal of Physical Oceanography, 1990, 20(2): 307−318. doi: 10.1175/1520-0485(1990)020<0307:OOTPNE>2.0.CO;2 [32] Hunt B, Strugnell J, Bednarsek N, et al. Poles apart: the “bipolar” pteropod species Limacina helicina is genetically distinct between the Arctic and Antarctic oceans[J]. PLoS One, 2010, 5(3): e9835. doi: 10.1371/journal.pone.0009835 [33] Hebert P D N, Stoeckle M Y, Zemlak T S, et al. Identification of birds through DNA barcodes[J]. PLoS Biology, 2004, 2(10): e312. doi: 10.1371/journal.pbio.0020312 [34] 李琪, 刘君, 孔令锋. 种的概念及种的界定与鉴定[J]. 中国海洋大学学报, 2014, 44(10): 57−64.Li Qi, Liu Jun, Kong Lingfeng. Species concept, species delimitation and species identification[J]. Periodical of Ocean University of China, 2014, 44(10): 57−64. [35] 林森杰, 王路, 郑连明, 等. 海洋生物DNA条形码研究现状与展望[J]. 海洋学报, 2014, 36(12): 1−17.Lin Senjie, Wang Lu, Zheng Lianming, et al. Current status and future prospect of DNA barcoding in marine biology[J]. Haiyang Xuebao, 2014, 36(12): 1−17. [36] Avise J C. Phylogeography: The History and Formation of Species[M]. Cambridge (Massachusetts): Harvard University Press, 2000: 447. [37] Avise J C. Molecular markers, natural history and evolution[M]. 2nd ed. Sunderland (Massachusetts): Sinauer Associates, 2004: 541. [38] Wiemers M, Fiedler K. Does the DNA barcoding gap exist?-a case study in blue butterflies (Lepidoptera: Lycaenidae)[J]. Frontiers in Zoology, 2007, 4: 8. doi: 10.1186/1742-9994-4-8 [39] Ortman B D, Bucklin A, Pagès F, et al. DNA barcoding the Medusozoa using mtCOI[J]. Deep Sea Research Part II: Topical Studies in Oceanography, 2010, 57(24/26): 2418−2156. [40] 刘青青, 董志军. 基于线粒体COI基因分析钩手水母的群体遗传结构[J]. 生物多样性, 2018, 26(11): 1204−1211. doi: 10.17520/biods.2018044Liu Qingqing, Dong Zhijun. Population genetic structure of Gonionemus vertens based on the mitochondrial COI sequence[J]. Biodiversity Science, 2018, 26(11): 1204−1211. doi: 10.17520/biods.2018044 [41] Bucklin A, Wiebe P H, Smolenack S B, et al. DNA barcodes for species identification of euphausiids (Euphausiacea, Crustacea)[J]. Journal of Plankton Research, 2007, 29(6): 483−493. doi: 10.1093/plankt/fbm031 [42] Durbin A, Hebert P D N, Cristescu M E A. Comparative phylogeography of marine cladocerans[J]. Marine Biology, 2008, 155(1): 1−10. doi: 10.1007/s00227-008-0996-x [43] Radulovici A E, Sainte-marie B, Dufresne F. DNA barcoding of marine crustaceans from the estuary and gulf of St Lawrence: a regional-scale approach[J]. Molecular Ecology Resources, 2009, 9(S1): 181−187. [44] Marlétaz F, Le Parco Y, Liu Shenglin, et al. Extreme mitogenomic variation in natural populations of chaetognaths[J]. Genome Biology and Evolution, 2017, 9(6): 1374−1384. doi: 10.1093/gbe/evx090 [45] Hubert N, Meyer C P, Bruggemann H J, et al. Cryptic diversity in indo-pacific coral-reef fishes revealed by DNA-barcoding provides new support to the Centre-of-overlap hypothesis[J]. PLoS One, 2012, 7(3): e28987. doi: 10.1371/journal.pone.0028987 -





 下载:
下载: