Effect of the content of colanic acid in marine bacterial biofilms on the settlement of Mytilus coruscus plantigrades
-
摘要: 可拉酸是生物被膜上重要的胞外多糖之一,但细菌可拉酸对海洋无脊椎动物附着过程的影响还鲜少研究。本研究从自然生物被膜中分离出8株海洋细菌,对其种属进行鉴定及聚类分析,并测定其生物被膜的可拉酸含量及对稚贝附着的诱导能力。筛选所得海洋细菌形成生物被膜并测定其成膜能力及胞外产物含量,发现β-多糖的生物量与厚壳贻贝稚贝附着率呈显著正相关趋势(p < 0.05)。8株海洋细菌生物被膜中可拉酸含量的定量结果显示,3株革兰氏阳性菌无法产生可拉酸,5株革兰氏阴性菌均可检测到不同含量的可拉酸,其中革兰氏阴性菌Shewanella marisflavi的可拉酸含量最高,为1 076.43 μg/mL。不同可拉酸含量的海洋细菌单一生物被膜与厚壳贻贝稚贝附着率之间关系的研究结果显示,海洋细菌生物被膜对厚壳贻贝稚贝附着率的诱导效果与其可拉酸含量呈显著正相关(p < 0.05)。以上结果表明,细菌生物被膜中的可拉酸能够参与诱导厚壳贻贝稚贝的附着。本研究为探究海洋细菌生物被膜的化学物质与海洋贝类附着之间的相互作用及其对贝类附着机制提出了新的见解。Abstract: Colanic acid is one of the vital exopolysaccharides in biofilms, yet the effect of marine bacterial colanic acid on the settlement of invertebrates is still rarely covered. In this study, eight strains of marine bacteria isolated from natural biofilms were identified and the phylogenetic analysis was carried out, the colanic acid content and inducing ability of biofilms were also determined. Before that, the biofilm formation capacity and the extracellular products of the screened bacteria were detected. It was found that β-polysaccharide had a significant positive correlation with the settlement rate of Mytilus coruscus plantigrades (p < 0.05). The quantitative results of colanic acid content showed that among the five Gram-negative bacteria which could produce colanic acid, Shewanella marisflavi had the highest colanic acid yield of 1076.43 μg/mL, and three Gram-positive bacteria could not generate it. The results of plantigrade settlement induced by different biofilms showed that the inducing activity of marine bacterial biofilms on the settlement rate of M. coruscus was positively correlated with the content of colanic acid (p < 0.05). These findings prove that the Gram-negative bacteria with colanic acid can positively regulate the settlement of M. coruscus plantigrades, while the Gram-positive bacteria without colanic acid have no inducing activity. This study provides further insights to figure out the interaction between the chemical cues of biofilms and marine molluscs.
-
Key words:
- biofilm /
- Mytilus coruscus /
- plantigrade /
- colanic acid /
- settlement
-
图 1 8株细菌的菌落形态及邻接法构建的系统发育树
a. 8株细菌的表型特征; b. 邻接法构建16S rDNA基因序列系统发育树
Fig. 1 The colony morphological and phylogenetic tree constructed by the Neighbor-Joining method of the eight marine bacterial strains
a. The morphological characteristics of eight bacterial species; b. phylogenetic tree of 16S rDNA gene sequences constructed by the Neighbor-Joining method
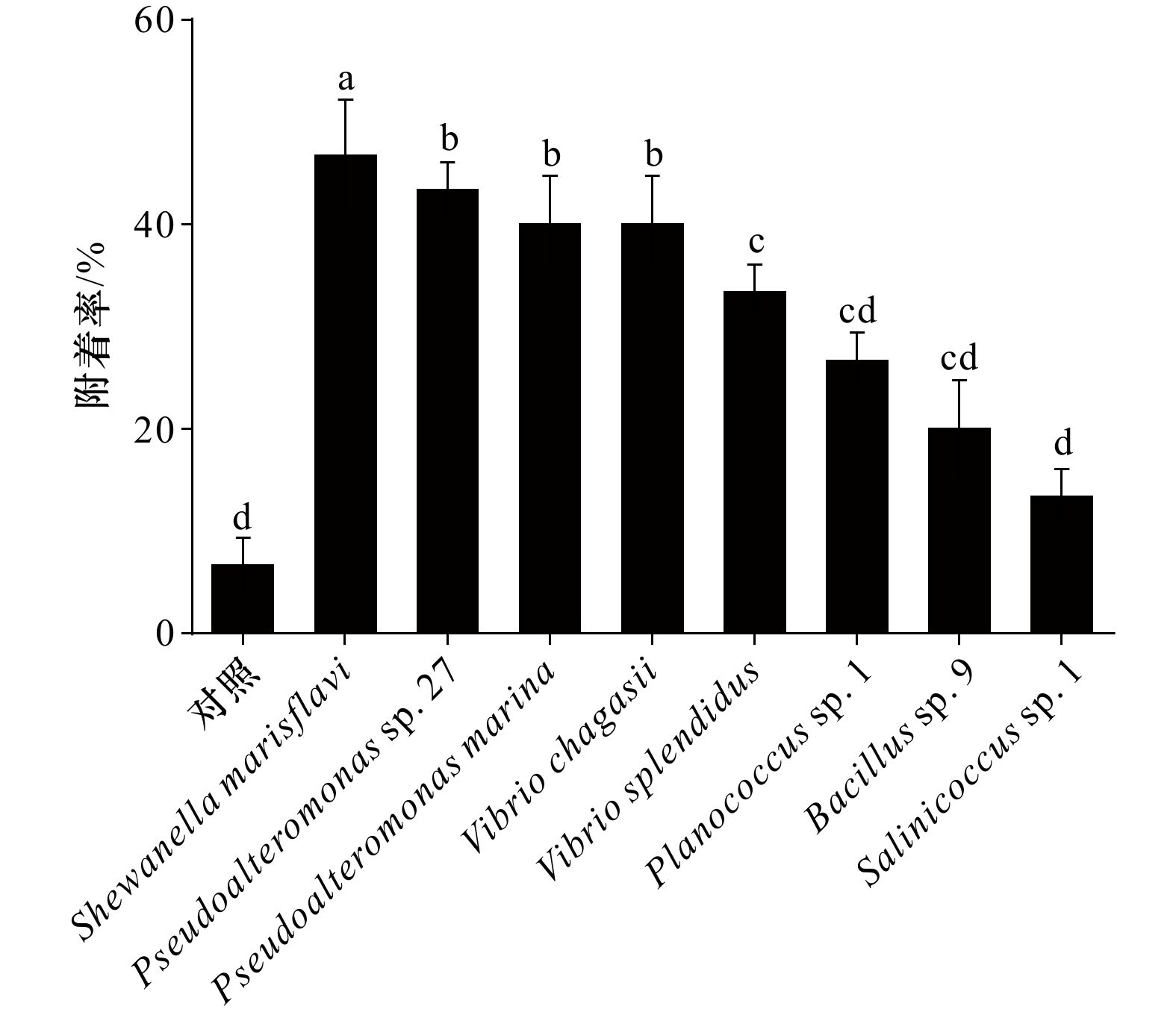
图 3 不同海洋细菌生物被膜形成能力
A. 不同海洋细菌形成生物被膜CLSM 拍摄图像;B. 不同海洋细菌形成生物被膜的终密度;C. 不同海洋细菌生物被膜膜厚;不同字母代表显著性差异
Fig. 3 Biofilm formation ability of different marine bacteria
A. Confocal laser scanning micrographs of biofilms; B. the density of eight monospecific bacterial biofilms; C. the thickness of biofilms; difference letters represent significant difference
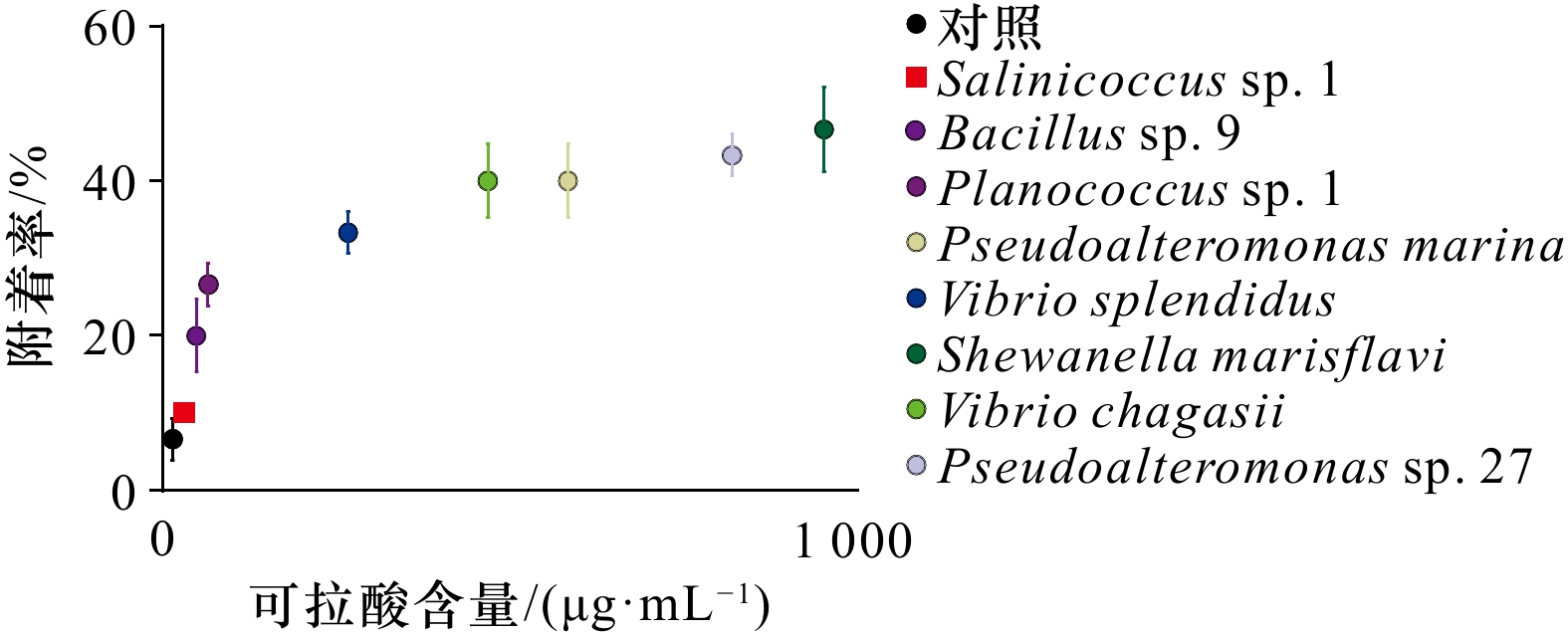
图 5 8株海洋细菌的可拉酸染色(A)及可拉酸含量测定(B)
A. 光镜下观察可拉酸在媒染生物膜上的分布,细菌被染成红色,可拉酸被染成蓝色;B. 生物被膜可拉酸浓度,红色箭头表示未检测到可拉酸,不同字母代表显著性差异
Fig. 5 The dyeing (A) and determination of the content (B) of colanic acid of eight marine pacteria
A. The distribution of colanic acid on mordant-stained biofilms under light microscope, the strains were stained red and the colanic acid was stained blue; B. the colanic acid concentration of biofilms, the red arrow indicates no colanic acid was detected, difference letters represent significant difference
表 1 激光扫描共聚焦显微镜荧光染料信息
Tab. 1 Confocal laser scanning microscope fluorescence dye information
染料 结合物质 波长/nm 碘化丙啶(PI) 死细菌 561 四甲基罗丹明共轭物刀豆蛋白( ConA-TMR) α-多糖 552/578 卡尔科弗卢尔荧光增白剂(CFW) β-多糖 254/432 1,1'−双十八烷基−3,3,3',3'− 四甲基吲哚二碳
花菁高氯酸盐(DiD’oil)脂质 648/670 异硫氰酸荧光素(FITC) 蛋白质 495/519 表 2 不同海洋细菌的16S rDNA序列分析
Tab. 2 16S rDNA sequence analysis of different marine bacteria
序列号 登录号 对照菌名 比对序列号 相似度/% 来源 ECSMB14101 CP041153 Shewanella marisflavi CP041153 99.58 自然生物被膜 ECSMB14103 CP023558 Pseudoalteromonas marina CP023558 99.79 自然生物被膜 ECSMB14105 MT820512 Vibrio splendidus MT820512 99.93 自然生物被膜 ECSMB14107 CP034970 Vibrio chagasii MN938232 99.24 自然生物被膜 ECSMB20101 OP209745 Planococcus sp. 1 MH938046 99.93 自然生物被膜 ECSMB20104 OP209750 Pseudoalteromonas sp. 27 KX889969 99.93 自然生物被膜 ECSMB20107 OP209748 Bacillus sp. 9 MG309326 99.86 自然生物被膜 ECSMB21103 OP209747 Salinicoccus sp. 1 DQ001316 98.22 自然生物被膜 表 3 8株海洋细菌遗传距离
Tab. 3 Genetic distance of the eight marine bacterial strains
测试菌株 Pseudoalteromonas
marinaPlanococcus
sp. 1Vibrio
splendidusVibrio
chagasiiSalinicoccus
sp. 1Bacillus
sp. 9Shewanella
marisflaviPseudoalteromonas
sp. 27Pseudoalteromonas marina 0 Planococcus sp. 1 0.262 0 Vibrio splendidus 0.126 0.259 0 Vibrio chagasii 0.139 0.267 0.036 0 Salinicoccus sp. 1 0.262 0.109 0.250 0.257 0 Bacillus sp. 9 0.256 0.088 0.263 0.261 0.113 0 Shewanella marisflavi 0.099 0.267 0.140 0.144 0.254 0.259 0 Pseudoalteromonas sp. 27 0.021 0.262 0.119 0.132 0.259 0.256 0.093 0 表 4 胞外产物与厚壳贻贝稚贝附着率相关性分析
Tab. 4 Correlation analyses between biofilm extracellular products and settlement rate of Mytilus coruscus plantigrades
胞外产物 附着率 r p α-多糖 0.584 0.129 β-多糖 0.742 0.035* 蛋白质 −0.656 0.077 脂质 0.579 0.132 注:p为检验值;r为相关系数;*表示显著性差异(p < 0.05)。 -
[1] 常亚青. 贝类增养殖学[M]. 北京: 中国农业出版社, 2007.Chang Yaqing. Mollusc Culture[M]. Beijing: China Agriculture Press, 2007. [2] 常抗美, 刘慧慧, 李家乐, 等. 紫贻贝和厚壳贻贝杂交及F1代杂交优势初探[J]. 水产学报, 2008, 32(4): 552−557.Chang Kangmei, Liu Huihui, Li Jiale, et al. A primary study on hybridization of Mytilus galloprovincialis, Mytilus coruscus, heterosis of F1 generation[J]. Journal of Fisheries of China, 2008, 32(4): 552−557. [3] Castro P, Huber M E. Marine Biology[M]. 10th ed. New York: McGraw-Hill Education, 2016. [4] 杨金龙, 郭行磐, 陈芋如, 等. 中湿度表面的海洋细菌对厚壳贻贝稚贝附着的影响[J]. 水产学报, 2015, 39(3): 421−428.Yang Jinlong, Guo Xingpan, Chen Yuru, et al. Effects of bacterial biofilms formed on middle wettability surfaces on settlement of plantigrades of the mussel Mytilus coruscus[J]. Journal of Fisheries of China, 2015, 39(3): 421−428. [5] 李慷均, 顾成柏. 厚壳贻贝的北方人工繁育技术[J]. 水产养殖, 2015, 36(8): 35−37.Li Kangjun, Gu Chengbai. Artificial breeding techniques of Mytilus coruscus in northern China. [J]. Journal of Aquaculture, 2015, 36(8): 35−37. [6] Yang Jinlong, Li Xiang, Liang Xiao, et al. Effects of natural biofilms on settlement of plantigrades of the mussel Mytilus coruscus[J]. Aquaculture, 2014, 424−425: 228−233. doi: 10.1016/j.aquaculture.2014.01.007 [7] Li Yifeng, Guo Xingpan, Yang Jinlong, et al. Effects of bacterial biofilms on settlement of plantigrades of the mussel Mytilus coruscus[J]. Aquaculture, 2014, 433: 434−441. doi: 10.1016/j.aquaculture.2014.06.031 [8] Li Yifeng, Chen Yuru, Yang Jinlong, et al. Effects of substratum type on bacterial community structure in biofilms in relation to settlement of plantigrades of the mussel Mytilus coruscus[J]. International Biodeterioration & Biodegradation, 2014, 96: 41−49. [9] Limoli D H, Jones C J, Wozniak D J. Bacterial extracellular polysaccharides in biofilm formation and function[J]. Microbiology Spectrum, 2015, 3(3): 223−247. [10] Sánchez-Lazo C, Martínez-Pita I. Biochemical and energy dynamics during larval development of the mussel Mytilus galloprovincialis (Lamarck, 1819)[J]. Aquaculture, 2012, 358−359: 71−78. doi: 10.1016/j.aquaculture.2012.06.021 [11] 梁箫, 刘红雨, 杨丽婷, 等. 弧菌生物被膜的动态演替对厚壳贻贝附着的影响[J]. 水产学报, 2020, 44(1): 118−129.Liang Xiao, Liu Hongyu, Yang Liting, et al. Effects of dynamic succession of Vibrio biofilms on settlement of the mussel Mytilus coruscus[J]. Journal of Fisheries of China, 2020, 44(1): 118−129. [12] Zeng Zhenshun, Guo Xingpan, Li Baiyuan, et al. Characterization of self-generated variants in Pseudoalteromonas lipolytica biofilm with increased antifouling activities[J]. Applied Microbiology and Biotechnology, 2015, 99(23): 10127−10139. doi: 10.1007/s00253-015-6865-x [13] Peng Lihua, Liang Xiao, Chang Ruiheng, et al. A bacterial polysaccharide biosynthesis-related gene inversely regulates larval settlement and metamorphosis of Mytilus coruscus[J]. Biofouling, 2020, 36(7): 753−765. doi: 10.1080/08927014.2020.1807520 [14] Liang Xiao, Zhang Junbo, Shao Anqi, et al. Bacterial cellulose synthesis gene regulates cellular c-di-GMP that control biofilm formation and mussel larval settlement[J]. International Biodeterioration & Biodegradation, 2021, 165: 105330. [15] Peng Lihua, Liang Xiao, Xu Jiakang, et al. Monospecific biofilms of Pseudoalteromonas promote larval settlement and metamorphosis of Mytilus coruscus[J]. Scientific Reports, 2020, 10(1): 2577. doi: 10.1038/s41598-020-59506-1 [16] Goebel W F. Colanic acid[J]. Proceedings of the National Academy of Sciences of the United States of America, 1963, 49(4): 464−471. [17] Stevenson G, Andrianopoulos K, Hobbs M, et al. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid[J]. Journal of Bacteriology, 1996, 178(16): 4885−4893. doi: 10.1128/jb.178.16.4885-4893.1996 [18] Grant W D, Sutherland I W, Wilkinson J F. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae[J]. Journal of Bacteriology, 1969, 100(3): 1187−1193. doi: 10.1128/jb.100.3.1187-1193.1969 [19] Shugar S, Sanderson K E. Characterization of a mucoid Escherichia coli K12 strain, and chemical analysis of the exopolysaccharide[J]. Canadian Journal of Microbiology, 1972, 18(7): 969−973. doi: 10.1139/m72-150 [20] Yu Min, Xu Ying, Xu Tingting, et al. WcaJ, the initiating enzyme for colanic acid synthesis, is required for lipopolysaccharide production, biofilm formation and virulence in Edwardsiella tarda[J]. Aquaculture, 2015, 437: 287−291. doi: 10.1016/j.aquaculture.2014.12.011 [21] Steenackers H, Hermans K, Vanderleyden J, et al. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication[J]. Food Research International, 2012, 45(2): 502−531. doi: 10.1016/j.foodres.2011.01.038 [22] Raiger Iustman L J, Tribelli P M, Ibarra J G, et al. Genome sequence analysis of Pseudomonas extremaustralis provides new insights into environmental adaptability and extreme conditions resistance[J]. Extremophiles, 2015, 19(1): 207−220. doi: 10.1007/s00792-014-0700-7 [23] Zdorovenko E L, Kadykova A A, Shashkov A S, et al. Pantoea agglomerans P1a lipopolysaccharide: structure of the O-specific polysaccharide and lipid A and biological activity[J]. Carbohydrate Research, 2019, 484: 107767. doi: 10.1016/j.carres.2019.107767 [24] 梁箫, 童欢, 彭莉华, 等. 纤维素对海洋细菌生物被膜形成及厚壳贻贝幼虫附着变态的调控[J]. 大连海洋大学学报, 2020, 35(1): 75−82. doi: 10.16535/j.cnki.dlhyxb.2019-025Liang Xiao, Tong Huan, Peng Lihua, et al. Regulation of formation of biofilms and larval settlement and metamorphosis of mussel Mytilus coruscus by cellulose[J]. Journal of Dalian Ocean University, 2020, 35(1): 75−82. doi: 10.16535/j.cnki.dlhyxb.2019-025 [25] Chen Jinru, Lee S M, Mao Ying. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157: H7 to osmotic and oxidative stress[J]. International Journal of Food Microbiology, 2004, 93(3): 281−286. doi: 10.1016/j.ijfoodmicro.2003.12.004 [26] Wang T K F, Yam W C, Yuen K Y, et al. Misidentification of a mucoid strain of Salmonella enterica serotype choleraesuis as Hafnia alvei by the Vitek GNI+ card system[J]. Journal of Clinical Microbiology, 2006, 44(12): 4605−4608. doi: 10.1128/JCM.01488-06 [27] Wang Chenhui, Zhang Hailing, Wang Jianli, et al. Colanic acid biosynthesis in Escherichia coli is dependent on lipopolysaccharide structure and glucose availability[J]. Microbiological Research, 2020, 239: 126527. doi: 10.1016/j.micres.2020.126527 [28] Yang Jinlong, Shen Peijing, Liang Xiao, et al. Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to monospecific bacterial biofilms[J]. Biofouling, 2013, 29(3): 247−259. doi: 10.1080/08927014.2013.764412 [29] Bao Weiyang, Yang Jinlong, Satuito C G, et al. Larval metamorphosis of the mussel Mytilus galloprovincialis in response to Alteromonas sp. 1: evidence for two chemical cues?[J]. Marine Biology, 2007, 152(3): 657−666. doi: 10.1007/s00227-007-0720-2 [30] González-Machado C, Capita R, Riesco-Peláez F, et al. Visualization and quantification of the cellular and extracellular components of Salmonella Agona biofilms at different stages of development[J]. PLoS One, 2018, 13(7): e0200011. doi: 10.1371/journal.pone.0200011 [31] Ren Ge, Wang Zhou, Li Ye, et al. Effects of lipopolysaccharide core sugar deficiency on colanic acid biosynthesis in Escherichia coli[J]. Journal of Bacteriology, 2016, 198(11): 1576−1584. doi: 10.1128/JB.00094-16 [32] Obadia B, Lacour S, Doublet P, et al. Influence of tyrosine-kinase Wzc activity on colanic acid production in Escherichia coli K12 cells[J]. Journal of Molecular Biology, 2007, 367(1): 42−53. doi: 10.1016/j.jmb.2006.12.048 [33] Yang Jinlong, Li Yifeng, Liang Xiao, et al. Silver nanoparticles impact biofilm communities and mussel settlement[J]. Scientific Reports, 2016, 6: 37406. doi: 10.1038/srep37406 [34] Weiner R M, Segall A M, Colwell R R. Characterization of a marine bacterium associated with Crassostrea virginica (the eastern oyster)[J]. Applied and Environmental Microbiology, 1985, 49(1): 83−90. doi: 10.1128/aem.49.1.83-90.1985 [35] Lau S C K, Mak K K W, Chen Feng, et al. Bioactivity of bacterial strains isolated from marine biofilms in Hong Kong waters for the induction of larval settlement in the marine polychaete Hydroides elegans[J]. Marine Ecology Progress Series, 2002, 226: 301−310. doi: 10.3354/meps226301 [36] 周轩, 郭行磐, 陈芋如, 等. 低湿度表面的海洋附着细菌对厚壳贻贝附着的影响[J]. 大连海洋大学学报, 2015, 30(1): 30−35. doi: 10.3969/J.ISSN.2095-1388.2015.01.006Zhou Xuan, Guo Xingpan, Chen Yuru, et al. Effects of bacterial biofilms formed on low surface wettability on settlement of plantigrades of the mussel Mytilus coruscus[J]. Journal of Dalian Ocean University, 2015, 30(1): 30−35. doi: 10.3969/J.ISSN.2095-1388.2015.01.006 [37] Tran C, Hadfield M G. Larvae of Pocillopora damicornis (Anthozoa) settle and metamorphose in response to surface-biofilm bacteria[J]. Marine Ecology Progress Series, 2011, 433: 85−96. doi: 10.3354/meps09192 [38] Watnick P, Kolter R. Biofilm, city of microbes[J]. Journal of Bacteriology, 2000, 182(10): 2675−2679. doi: 10.1128/JB.182.10.2675-2679.2000 [39] Characklis W G, Cooksey K E. Biofilms and microbial fouling[J]. Advances in Applied Microbiology, 1983, 29: 93−138. [40] Flemming H C, Wingender J. The biofilm matrix[J]. Nature Reviews Microbiology, 2010, 8(9): 623−633. doi: 10.1038/nrmicro2415 [41] Seviour T, Derlon N, Dueholm M S, et al. Extracellular polymeric substances of biofilms: suffering from an identity crisis[J]. Water Research, 2019, 151: 1−7. doi: 10.1016/j.watres.2018.11.020 [42] Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli[J]. Annual Review of Biochemistry, 2006, 75: 39−68. doi: 10.1146/annurev.biochem.75.103004.142545 [43] Dawirs R R, Dietrich A. Temperature and laboratory feeding rates in Carcinusmaenas L. (Decapoda: Portunidae) larvae from hatching through metamorphosis[J]. Journal of Experimental Marine Biology and Ecology, 1986, 99(2): 133−147. doi: 10.1016/0022-0981(86)90233-9 [44] Chan D C. Mitochondria: dynamic organelles in disease, aging, and development[J]. Cell, 2006, 125(7): 1241−1252. doi: 10.1016/j.cell.2006.06.010 [45] Yao Yanhua, Tsuchiyama S, Yang Ciyu, et al. Proteasomes, Sir2, and Hxk2 form an interconnected aging network that impinges on the AMPK/Snf1-regulated transcriptional repressor Mig1[J]. PLoS Genetics, 2015, 11(1): e1004968. doi: 10.1371/journal.pgen.1004968 [46] Han Bing, Sivaramakrishnan P, Lin C C J, et al. Microbial genetic composition tunes host longevity[J]. Cell, 2017, 169(7): 1249−1262.e13. doi: 10.1016/j.cell.2017.05.036 [47] Li Zheng, Okamoto K I, Hayashi Y, et al. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses[J]. Cell, 2004, 119(6): 873−887. doi: 10.1016/j.cell.2004.11.003 -





 下载:
下载: