Virus-induced autophagy in the marine coccolithophorid Emiliania huxleyi
-
摘要: 以海洋环境中具有重要生态功能的赫氏颗石藻(Emiliania huxleyi)BOF92及其特异性裂解病毒(E. huxleyi virus, EhV99B1)为研究对象,采用分子及细胞生物学方法探究海洋病毒感染诱导宿主细胞自噬特征及其调控机制。结果显示:病毒感染能诱导宿主细胞自噬,并出现明显的液泡酸化及液泡自噬现象;感染过程中核心自噬相关基因如atg1、atg5、atg8及atg12的mRNA表达水平均显著上调(p < 0.01),自噬启动成核相关蛋白Vps34显著上调(p < 0.01),进而启动自噬并促使自噬体与液泡的融合;自噬标志性蛋白p62显著下调(p < 0.05),表明自噬流畅通以加速蛋白降解;感染中后期对自噬起负调控作用的磷脂酰肌醇(PI3K)、磷酸化蛋白激酶B(p-Akt)和磷酸化雷帕霉素靶蛋白(p-TOR)等因子的表达水平均显著降低(p < 0.01)。另外,病毒感染过程中,细胞活性氧(ROS)水平显著升高(p < 0.01),线粒体膜电位(MMP)及ATP含量显著降低(p < 0.01)。综上,病毒感染诱发宿主藻细胞ROS的产生、线粒体膜受损,并通过调节PI3K/Akt/TOR级联反应诱导细胞自噬。可见,自噬作为一种独特的程序性细胞死亡形式,对浮游植物遭受胁迫后的个体存活及种群延续具有重要意义。Abstract: To understand the characteristics of autophagy induced by virus infection in microalgae Emiliania huxleyi, we used diverse techniques including transmission electron microscopy, fluorescence microscopy, immunolabeling and biochemical methodologies to investigate the role of autophagy in the interaction between E. huxleyi BOF92 and its specific virus EhV99B1. The results showed that virus infection induced autophagy and vacuolar acidification in host cells, concomitant with up-regulation of autophagy-related genes such as atg1, atg5, atg8 and atg12 (p < 0.01) and Vps34 protein involved in the induction and nucleation of autophagosomes (p < 0.01). The expression level of autophagy marker protein p62 was significantly down-regulated (p < 0.05) during viral infection, indicating enhanced autophagic flux and activated autophagy. The expressions levels of negative regulatory factors such as phosphatidylinositol (PI3K), phosphorylated protein kinase B (p-Akt) and phosphorylated target of rapamycin protein (p-TOR) were significantly decreased in the late stage of viral infection (p < 0.01). Moreover, the level of reactive oxygen species (ROS) increased dramatically (p < 0.01), accompanied by a significant reduction in mitochondrial membrane potential (MMP) and ATP levels (p < 0.01) during viral infection. In conclusion, EhV99B1 infection induces ROS production and mitochondrial membrane damage in host cells, and initiates autophagy by regulating the PI3K/Akt/TOR signal pathway. Therefore, autophagy, as a unique form of programmed cell death, is of great significance to the individual survival and population dynamics of phytoplankton respond to environmental and biological stress.
-
Key words:
- Emiliania huexlyi /
- virus /
- autophagy /
- autophagy-related genes /
- mitochondrial function /
- PI3K/Akt/TOR signal pathway
-
图 1 EhV99B1感染诱导颗石藻(E. huxleyi) BOF92细胞的裂解及细胞超微结构变化
a−c. 病毒感染导致藻细胞裂解(a. 未添加病毒的对照组;b. 病毒感染24 h;c. 病毒感染48 h);d−i. 病毒感染过程中藻细胞超微结构的变化(d. 对照组;e−g. 病毒感染24 h;h−i. 病毒感染48 h);N表示细胞核;C表示叶绿体;M表示线粒体;V表示液泡;黑色箭头指向自噬体或病毒颗粒,图e−h右下角的方框为该图中箭头所指位置的局部放大图
Fig. 1 Infection dynamics and ultrastructure changes of E. huxleyi BOF92 during EhV99B1 infection
a−c. Virus infection leads to algae cell lysis (a. uninfected culture; b. 24 h post infection; c. 48 h post infection); d−i. transmission electron micrograph of E. huxleyi cells during viral infection (d. uninfected cells; e−g. 24 h post infection; h−i. 48 h post infection); N: nucleus; C: chloroplast; M: mitochondrial; V: vacuole; black arrows point to autophagosome or virions, the insets in Fig. e−h show higher magnification of the boxed areas, depicting the double membrane vesicles
图 2 颗石藻细胞中酸化区室的Lysosensor荧光标记结果
对照组:未添加病毒;感染组:病毒感染48 h;雷帕霉素组:阳性对照,采用终浓度为10 μmol/L的雷帕霉素处理藻细胞48 h
Fig. 2 Results of Lysosensor fluorescent labeling of acidizing compartments in Emiliania huxleyi cells
Control group: uninfected cells; infected group: viral infected cells 48 h post infection; rapamycin group: 10 μmol/L rapamycin-treated cells for 48 h (positive control)
图 3 病毒感染对颗石藻细胞核心自噬相关基因表达水平的影响
a. 核心自噬相关基因qRT-PCR结果聚类热图(数值为感染组与对照组的比值),Con 6-1表示对照组1 6 h后基因表达水平,Inf 6-1表示感染组1 6 h后基因表达水平,以此类推;b. 自噬相关基因相对表达量;*表示差异显著(p < 0.05),**表示差异较显著(p < 0.01),***表示差异极显著(p < 0.001)
Fig. 3 Relative expression level effect of core autophagy-related genes in E. huxleyi cells during viral infection
a. qRT-PCR analysis profiles of autophagy related genes,the value is the ratio of infected group to control group. Con 6-1 represents the gene expression level of the control group 1 after 6 h, Inf 6-1 represents the gene expression level of the infected group 1 after 6 h, and so on; b. relative expression levels of selected autophagy related genes; * indicates a significant differ(p < 0.05), ** indicates more significant differ (p < 0.01), *** indicates an extreme differ (p < 0.001)
图 4 病毒感染对颗石藻细胞核心自噬相关蛋白表达的影响
*表示差异显著(p < 0.05),**表示差异较显著(p < 0.01);***表示差异极显著(p < 0.001)
Fig. 4 Expression of core autophagy-related proteins in E. huxleyi cells during viral infection
* Indicates a significant differ (p < 0.05), ** indicates more significant differ (p < 0.01), ***indicates an extreme differ (p < 0.001)
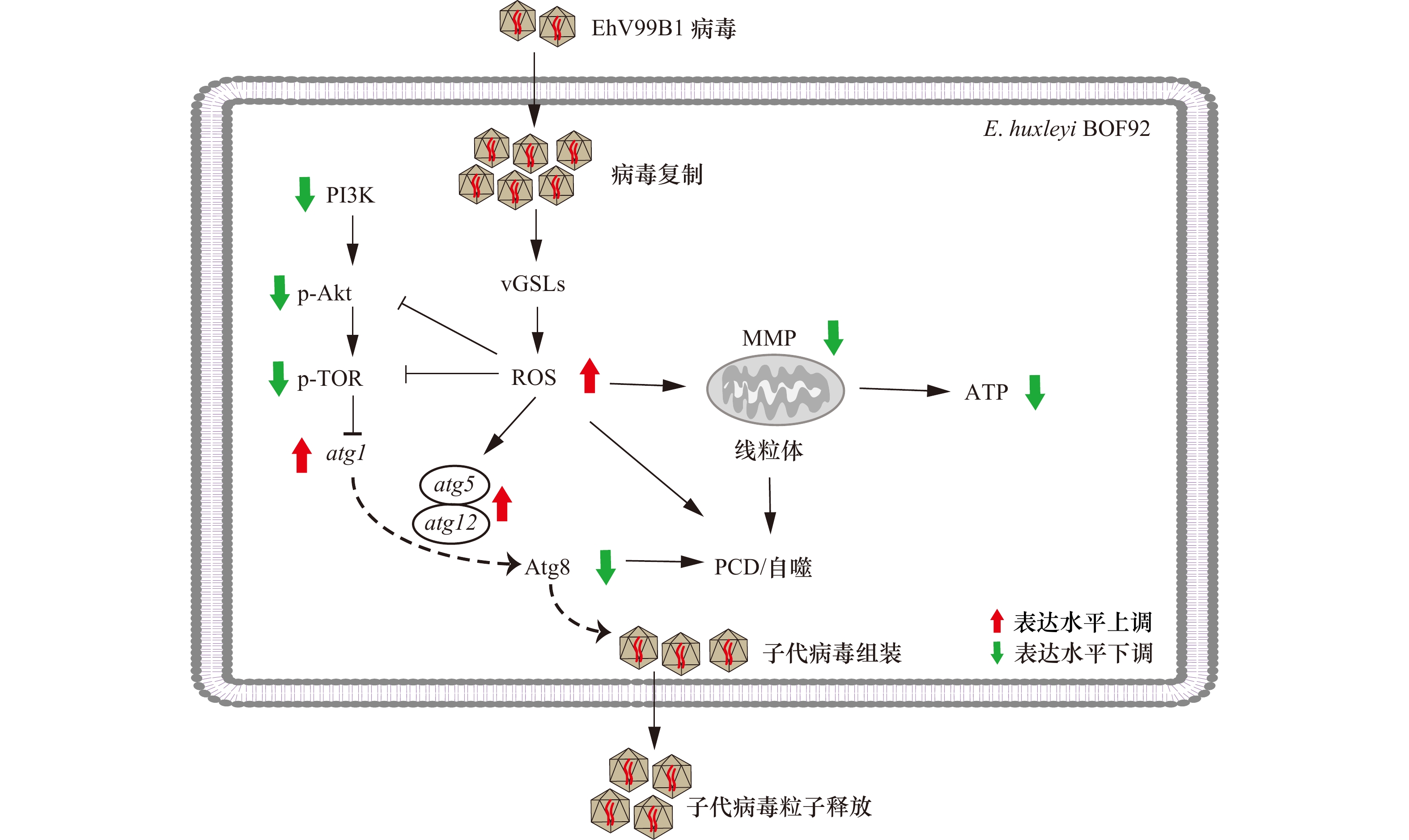
图 8 病毒感染诱导颗石藻细胞自噬示意图
图中粗箭头所示为atg1、atg5及atg12的mRNA表达水平、Atg8、PI3K、p-Akt及p-TOR的蛋白表达水平和ROS、MMP和ATP含量的变化
Fig. 8 Schematic diagram of virus induced autophagy in the E. huxleyi-EhV system
Thick arrows indicate the mRNA expression level of atg1, atg5, atg12, protein expression levels of Atg8, PI3K, p-Akt, p-TOR and contents of ATP, ROS, MMP
表 1 本研究自噬相关基因qRT-PCR引物序列
Tab. 1 Primer sequence of autophagy-related gene for qRT-PCR used in this study
基因
名称扩增片段
长度/bp引物
名称序列(5’→3’) atg1 106 F AGGGCAGCTTTGCGATTGT R TTGGCCTGTAGCTTCTGGTTG atg2 96 F CATGGGCGTGTTTGTCGAG R AAGGCGTAGAGGGAGGAGAAG atg4_1 118 F GCGTGCCTTTGGTTCACGTA R GTGCTTGCGCGAGTATCATCT atg3 146 F TCCTGGAGAAGGGTGTGTTGAC R CGTGACGAGGTACTGCTTGTTC atg5 117 F GAGCACTTCCTGCCTTTTGC R CGTCAGCAGGTCAAAGAGCA atg7 154 F GCAAACACACTCGAGGACTTCAA R TCTTGAGGTCTGCGAAGGTGA atg8_1 163 F GCCGCTGATTGACAAGAAGAA R CGTCCTTGTGGCTGTCGTAGA atg9 215 F GTCGCCCGTTTCGTCACTT R CGAGCCATCACAGCAGGTT atg10 181 F GGTGCTGGAGGAGCCAATCT R AGCGGCAGATGTGTATGCG atg12 149 F CGAACCGCTGCTCCTCTACT R GAAAGCACTGCGCCACATC atg13 249 F GCGAAACTGCGTCCAGAAGA R CGCTCGAGAAGCACGAGATG LST8 212 F TGGCAAACAACTTCTCGTGC R TCAAACTGGCCCATCGTGTA TOR_1 110 F CAGCACAATCTCCTTCTGCAC R TGCCGATTTGCGGGTAC TOR_3 79 F GACCGGCACCTCAACAACA R CCTCGAAGCAGTCGCCATA Vps15 122 F CATCAAGGGCGAGAATGTGC R CGTCAAAGTAGAAGGAGAAGTCCG Vps34_2 203 F AGCTCGTCTGGAAGTTCCGGTA R CCTCGGCTGTATCTCGCATACAT β-tubulin 160 F TCATGTGCTCCTACTCGGTCTTC R TTCAGCGTGCGGAAACAGA 表 2 本研究中使用的抗体
Tab. 2 Antibodies used in this study
抗体名称 生产商 产品货号 稀释倍数 CDK1 Beyotime, 中国 AF1516 1∶800 ATG8 Abcam, 英国 ab4753 1∶1000 Vps34 Abcam, 英国 ab233437 1∶200 CDK1 Beyotime, 中国 AF0111 1∶1000 PI3K Beyotime, 中国 AF7749 1∶1000 p-AKT (Thr308) Beyotime, 中国 AF5734 1∶1000 p-TOR (Ser2448) Bioworld, 中国 BS4706 1∶1000 IgG Thermo, 美国 31460 1∶10000 -
[1] 张健, 李佳芮, 杨璐, 等. 球石藻及其生态功能[J]. 海洋科学, 2018, 42(2): 150−158.Zhang Jian, Li Jiarui, Yang Lu, et al. Coccolithophores and their characteristics[J]. Marine Sciences, 2018, 42(2): 150−158. [2] Daniels C J, Poulton A J, Balch W M, et al. A global compilation of coccolithophore calcification rates[J]. Earth System Science Data, 2018, 10(4): 1859−1876. doi: 10.5194/essd-10-1859-2018 [3] Jensen L Ø, Mousing E A, Richardson K. Using species distribution modelling to predict future distributions of phytoplankton: case study using species important for the biological pump[J]. Marine Ecology, 2017, 38(3): e12427. doi: 10.1111/maec.12427 [4] 孙军. 今生颗石藻的有机碳泵和碳酸盐反向泵[J]. 地球科学进展, 2007, 22(12): 1231−1239. doi: 10.3321/j.issn:1001-8166.2007.12.003Sun Jun. Organic carbon pump and carbonate counter pump of living coccolithophorid[J]. Advances in Earth Science, 2007, 22(12): 1231−1239. doi: 10.3321/j.issn:1001-8166.2007.12.003 [5] Liu Jingwen, Cai Weicong, Fang Xian, et al. Virus-induced apoptosis and phosphorylation form of metacaspase in the marine coccolithophorid Emiliania huxleyi[J]. Archives of Microbiology, 2018, 200(3): 413−422. doi: 10.1007/s00203-017-1460-4 [6] Bidle K D, Haramaty L, Barcelos E Ramos J, et al. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi[J]. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(14): 6049−6054. [7] Kuhlisch C, Schleyer G, Shahaf N, et al. Viral infection of algal blooms leaves a unique metabolic footprint on the dissolved organic matter in the ocean[J]. Science Advances, 2021, 7(25): eabf4680. doi: 10.1126/sciadv.abf4680 [8] Bidle K D. The molecular ecophysiology of programmed cell death in marine phytoplankton[J]. Annual Review of Marine Science, 2015, 7: 341−375. doi: 10.1146/annurev-marine-010213-135014 [9] Bidle K D. Programmed cell death in unicellular phytoplankton[J]. Current Biology, 2016, 26(13): R594−R607. doi: 10.1016/j.cub.2016.05.056 [10] Yang Meng, Liu Yule. Autophagy in plant viral infection[J]. FEBS Letters, 2022, 596(17): 2152−2162. doi: 10.1002/1873-3468.14349 [11] 孙彦鹤, 周广舟, 王雨润, 等. 病毒诱导的细胞自噬与凋亡的串话关系研究进展[J]. 病毒学报, 2020, 36(4): 735−743.Sun Yanhe, Zhou Guangzhou, Wang Yurun, et al. Research progress in the crosstalk between the autophagy and apoptosis induced by viruses[J]. Chinese Journal of Virology, 2020, 36(4): 735−743. [12] 张杨, 孙弯弯, 陆丽丹, 等. 细胞自噬与凋亡相互作用分子机制的研究进展[J]. 基础医学与临床, 2021, 41(9): 1342−1346.Zhang Yang, Sun Wanwan, Lu Lidan, et al. Research progress on molecular mechanism of the interaction between autophagy and apoptosis[J]. Basic and Clinical Medicine, 2021, 41(9): 1342−1346. [13] Luo Chunshan, Liang Junrong, Lin Qun, et al. Cellular responses associated with ROS production and cell fate decision in early stress response to iron limitation in the diatom Thalassiosira pseudonana[J]. Journal of Proteome Research, 2014, 13(12): 5510−5523. doi: 10.1021/pr5004664 [14] Zeng Jun, Liu Sishangyu, Cai Weicong, et al. Emerging lipidome patterns associated with marine Emiliania huxleyi-virus model system[J]. Science of the Total Environment, 2019, 688: 521−528. doi: 10.1016/j.scitotenv.2019.06.284 [15] Vardi A, Van Mooy B A S, Fredricks H F, et al. Viral glycosphingolipids induce lytic infection and cell death in marine phytoplankton[J]. Science, 2009, 326(5954): 861−865. doi: 10.1126/science.1177322 [16] Bramucci A R, Case R J. Phaeobacter inhibens induces apoptosis-like programmed cell death in calcifying Emiliania huxleyi[J]. Scientific Reports, 2019, 9(1): 5215. doi: 10.1038/s41598-018-36847-6 [17] Pérez-Pérez M E, Couso I, Crespo J L. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii[J]. Autophagy, 2012, 8(3): 376−388. doi: 10.4161/auto.18864 [18] Pérez-Martín M, Blaby-Haas C E, Pérez-Pérez M E, et al. Activation of autophagy by metals in Chlamydomonas reinhardtii[J]. Eukaryotic Cell, 2015, 14(9): 964−973. doi: 10.1128/EC.00081-15 [19] Pugkaew W, Meetam M, Ponpuak M, et al. Role of autophagy in triacylglycerol biosynthesis in Chlamydomonas reinhardtii revealed by chemical inducer and inhibitors[J]. Journal of Applied Phycology, 2018, 30(1): 15−22. doi: 10.1007/s10811-017-1166-7 [20] Barreto Filho M M, Durand P M, Andolfato N E, et al. Programmed cell death in the coccoid green microalga Ankistrodesmus densus Korshikov (Sphaeropleales, Selenastraceae)[J]. European Journal of Phycology, 2022, 57(2): 193−206. doi: 10.1080/09670262.2021.1938240 [21] Shemi A, Schatz D, Fredricks H F, et al. Phosphorus starvation induces membrane remodeling and recycling in Emiliania huxleyi[J]. New Phytologist, 2016, 211(3): 886−898. doi: 10.1111/nph.13940 [22] Schatz D, Shemi A, Rosenwasser S, et al. Hijacking of an autophagy-like process is critical for the life cycle of a DNA virus infecting oceanic algal blooms[J]. New Phytologist, 2014, 204(4): 854−863. doi: 10.1111/nph.13008 [23] Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1[J]. Nature Cell Biology, 2011, 13(2): 132−141. doi: 10.1038/ncb2152 [24] 卢雪, 蔡伟聪, 刘静雯. 浮游植物细胞自噬作用特点及研究方法[J]. 微生物学杂志, 2020, 40(4): 105−113. doi: 10.3969/j.issn.1005-7021.2020.04.016Lu Xue, Cai Weicong, Liu Jingwen. Features and research methods summarization on phytoplankton cell autophagy[J]. Journal of Microbiology, 2020, 40(4): 105−113. doi: 10.3969/j.issn.1005-7021.2020.04.016 [25] Nissimov J I, Pagarete A, Ma Fangrui, et al. Coccolithoviruses: a review of cross-kingdom genomic thievery and metabolic thuggery[J]. Viruses, 2017, 9(3): 52. doi: 10.3390/v9030052 [26] Castberg T, Thyrhaug R, Larsen A, et al. Isolation and characterization of a virus that infects Emiliania huxleyi (Haptophyta)[J]. Journal of Phycology, 2002, 38(4): 767−774. doi: 10.1046/j.1529-8817.2002.02015.x [27] 隋馨莹, 徐平, 段昌柱, 等. p62蛋白的分子功能及其在疾病中的研究进展[J]. 生物工程学报, 2023, 39(4): 1374−1389.Sui Xinying, Xu Ping, Duan Changzhu, et al. Advances in molecular function of p62 protein and its role in diseases[J]. Chinese Journal of Biotechnology, 2023, 39(4): 1374−1389. [28] 刘洋, 张静, 王秋玲, 等. 植物细胞自噬研究进展[J]. 植物学报, 2018, 53(1): 5−16.Liu Yang, Zhang Jing, Wang Qiuling, et al. Research progress in plant autophagy[J]. Chinese Bulletin of Botany, 2018, 53(1): 5−16. [29] 黄晓, 李发强. 细胞自噬在植物细胞程序性死亡中的作用[J]. 植物学报, 2016, 51(6): 859−862.Huang Xiao, Li Faqiang. Roles of autophagy in plant programmed cell death[J]. Chinese Bulletin of Botany, 2016, 51(6): 859−862. [30] Minina E A, Filonova L H, Fukada K, et al. Autophagy and metacaspase determine the mode of cell death in plants[J]. Journal of Cell Biology, 2013, 203(6): 917−927. doi: 10.1083/jcb.201307082 [31] Pérez-Pérez M E, Couso I, Heredia-Martínez L G, et al. Monitoring autophagy in the model green microalga Chlamydomonas Reinhardtii[J]. Cells, 2017, 6(4): 36. doi: 10.3390/cells6040036 [32] Kirkegaard K, Taylor M P, Jackson W T. Cellular autophagy: surrender, avoidance and subversion by microorganisms[J]. Nature Reviews Microbiology, 2004, 2(4): 301−314. doi: 10.1038/nrmicro865 [33] Randow F, Münz C. Autophagy in the regulation of pathogen replication and adaptive immunity[J]. Trends in Immunology, 2012, 33(10): 475−487. doi: 10.1016/j.it.2012.06.003 [34] Wileman T. Aggresomes and autophagy generate sites for virus replication[J]. Science, 2006, 312(5775): 875−878. doi: 10.1126/science.1126766 [35] Fulton J M, Fredricks H F, Bidle K D, et al. Novel molecular determinants of viral susceptibility and resistance in the lipidome of Emiliania huxleyi[J]. Environmental Microbiology, 2014, 16(4): 1137−1149. doi: 10.1111/1462-2920.12358 [36] Kim J, Kim Y C, Fang Chong, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy[J]. Cell, 2013, 152(1/2): 290−303. [37] Lin Tianji, Ruan Shijuan, Huang Dingbang, et al. MeHg-induced autophagy via JNK/Vps34 complex pathway promotes autophagosome accumulation and neuronal cell death[J]. Cell Death & Disease, 2019, 10(6): 399. [38] Surviladze Z, Sterk R T, DeHaro S A, et al. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-Kinase/Akt/mTOR pathway and inhibition of autophagy[J]. Journal of Virology, 2013, 87(5): 2508−2517. doi: 10.1128/JVI.02319-12 [39] Tsapras P, Nezis I P. Caspase involvement in autophagy[J]. Cell Death and Differentiation, 2017, 24(8): 1369−1379. doi: 10.1038/cdd.2017.43 [40] 李杰, 丁奕, 项荣, 等. 浮游植物程序性细胞死亡研究进展[J]. 生态环境学报, 2010, 19(11): 2743−2748. doi: 10.3969/j.issn.1674-5906.2010.11.040Li Jie, Ding Yi, Xiang Rong, et al. Programmed cell death in phytoplankton[J]. Ecology and Environmental Sciences, 2010, 19(11): 2743−2748. doi: 10.3969/j.issn.1674-5906.2010.11.040 [41] Qian Miao, Tan Haiming, Yu Ning, et al. Inactivated Sendai virus induces ROS-dependent apoptosis and autophagy in human prostate cancer cells[J]. Biomedical and Environmental Sciences, 2018, 31(4): 280−289. [42] Zhang Yun, Yan Ming, Kuang Shumeng, et al. Bisphenol A induces apoptosis and autophagy in murine osteocytes MLO-Y4: involvement of ROS-mediated mTOR/ULK1 pathway[J]. Ecotoxicology and Environmental Safety, 2022, 230: 113119. doi: 10.1016/j.ecoenv.2021.113119 [43] Affenzeller M J, Darehshouri A, Andosch A, et al. PCD and autophagy in the unicellular green alga Micrasterias denticulata[J]. Autophagy, 2009, 5(6): 854−855. doi: 10.4161/auto.8791 [44] Sheyn U, Rosenwasser S, Ben-Dor S, et al. Modulation of host ROS metabolism is essential for viral infection of a bloom-forming coccolithophore in the ocean[J]. The ISME Journal, 2016, 10(7): 1742−1754. doi: 10.1038/ismej.2015.228 [45] 田雪, 蔡伟聪, 苏金净, 等. 病毒感染海洋球石藻 Emiliania huxleyi 的转录组分析[J]. 海洋学报, 2019, 41(12): 103−112.Tian Xue, Cai Weicong, Su Jinjing, et al. Transcriptome analysis of marine microalga Emiliania huxleyi in response to virus infection[J]. Haiyang Xuebao, 2019, 41(12): 103−112. [46] Zhang Enquan, Gao Jingjing, Wei Zehua, et al. MicroRNA-mediated regulation of lipid metabolism in virus-infected Emiliania huxleyi[J]. The ISME Journal, 2022, 16(11): 2457−2466. doi: 10.1038/s41396-022-01291-y [47] Kimmance S A, Allen M J, Pagarete A, et al. Reduction in photosystem II efficiency during a virus-controlled Emiliania huxleyi bloom[J]. Marine Ecology Progress Series, 2014, 495: 65−76. doi: 10.3354/meps10527 [48] 马丹颖, 季东超, 徐勇, 等. 活性氧调控植物细胞自噬的研究进展[J]. 植物学报, 2019, 54(1): 81−92. doi: 10.11983/CBB18012Ma Danying, Ji Dongchao, Xu Yong, et al. Advances in the regulation on autophagy by reactive oxygen species in plant cells[J]. Chinese Bulletin of Botany, 2019, 54(1): 81−92. doi: 10.11983/CBB18012 -





 下载:
下载: