Knockout of Pseudoalteromonas marina pilZ gene inhibited the settlement and metamorphosis of Mytilus coruscus
-
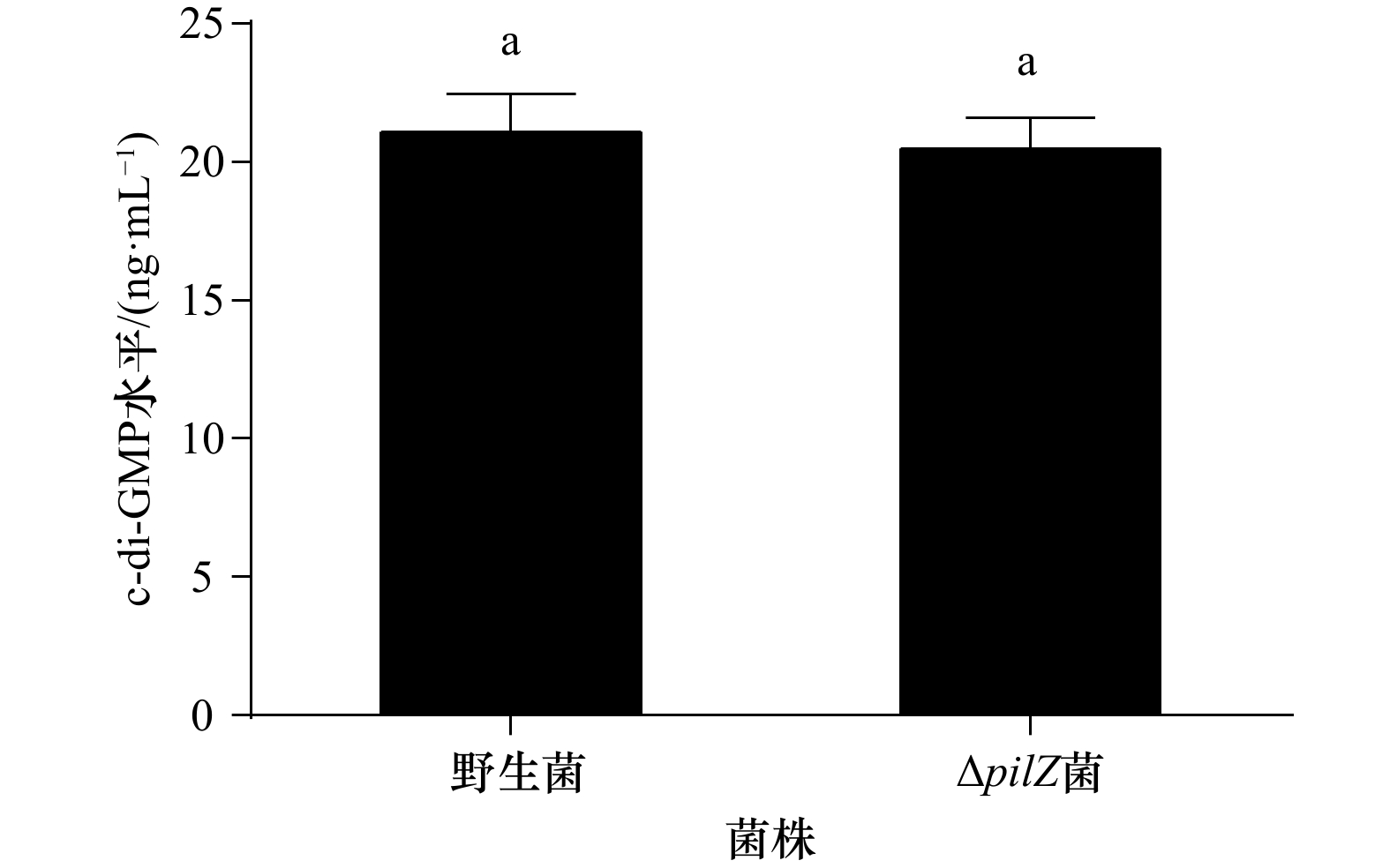
摘要: 为探究海假交替单胞菌pilZ基因的缺失对生物被膜形成及突变菌生物被膜对厚壳贻贝幼虫附着变态的影响,本文通过同源重组构建pilZ基因缺失菌,分析了基因缺失菌生物被膜的细菌密度、膜厚、c-di-GMP水平和胞外产物含量等特性的变化及其对厚壳贻贝幼虫附着变态的调控作用。结果表明:与野生型菌株相比,pilZ基因缺失菌形成的生物被膜膜厚增加、细菌数量增多,胞外产物中β-多糖、蛋白含量减少,抑制了厚壳贻贝幼虫的附着变态(p<0.05);而c-di-GMP水平、α-多糖和脂质含量无显著变化(p>0.05)。由此可见,海假交替单胞菌pilZ基因的缺失可调控细菌生物被膜形成和胞外产物包括β-多糖、蛋白质的含量,从而抑制厚壳贻贝幼虫的附着变态。Abstract: To investigate the effect of Pseudoalteromonas marina pilZ gene knockout on the biofilm formation and its influence on the settlement and metamorphosis of Mytilus coruscus larvae, ΔpilZ was constructed by homologous recombination, and the changes in bacterial density, biofilm thickness, c-di-GMP level and extracellular polymeric substances (EPS) of ΔpilZ bacteria biofilm were analyzed, as well as the regulation of the settlement and metamorphosis of M. coruscus larvae. The results showed that compared with the wild-type strain, the biofilm formed by ΔpilZ strain significantly increased the biofilm thickness, the number of bacteria, while the contents of β-polysaccharides and proteins in EPS were decreased and the settlement and metamorphosis of M. coruscus larvae were inhibited (p<0.05). There was no significant difference in c-di-GMP level, α-polysaccharide and lipid contents (p>0.05). In conclusion, P. marina pilZ gene knockout can regulate the bacterial biofilm and the content of EPS including β-polysaccharides and proteins contents, then inhibit the settlement and metamorphosis of M. coruscus larvae.
-
Key words:
- pilZ /
- biofilm /
- Mytilus coruscus /
- settlement and metamorphosis /
- extracellular polymeric substances
-
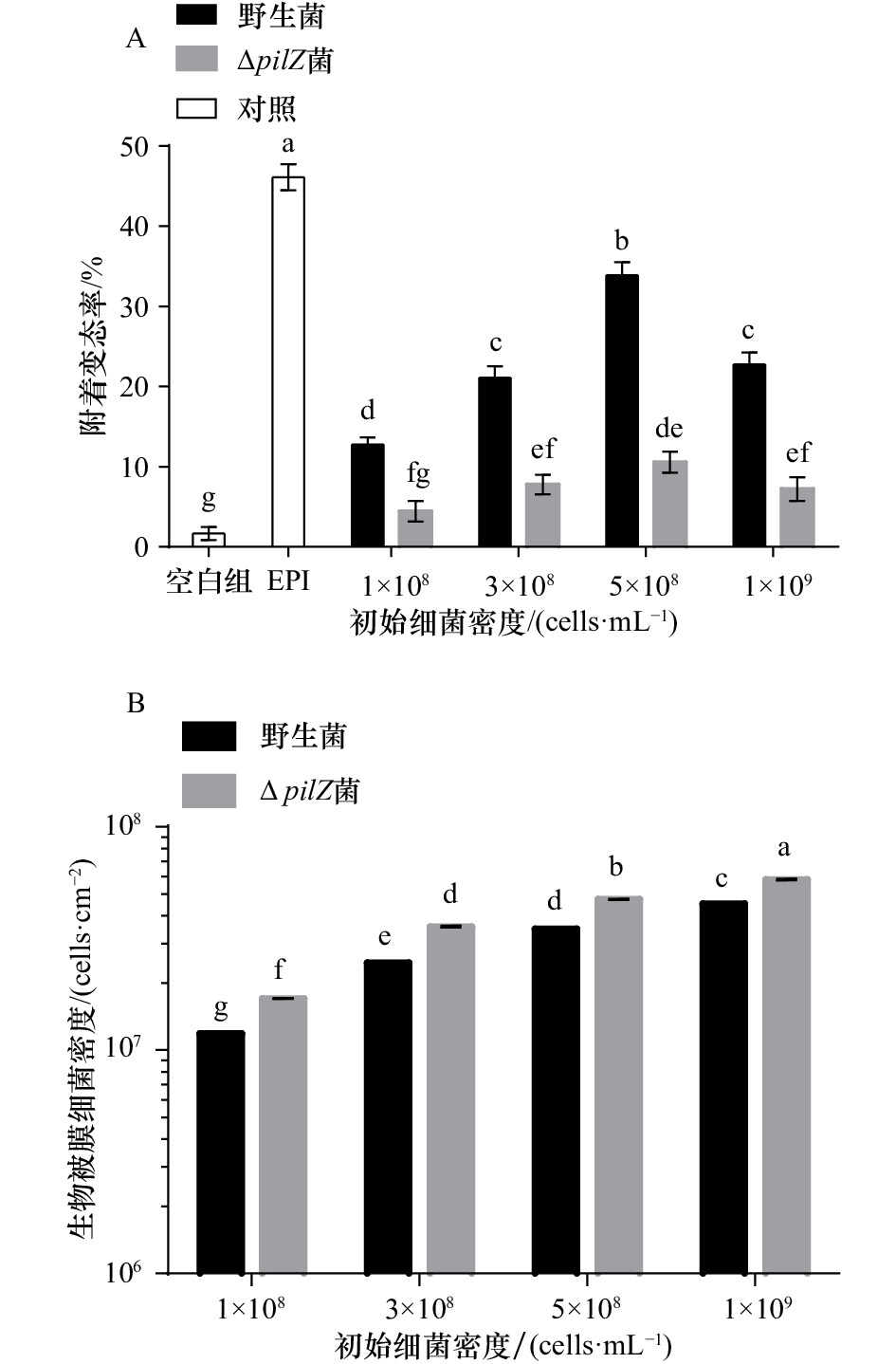
图 2 不同初始细菌密度下野生菌与ΔpilZ菌生物被膜对幼虫附着变态的影响(A)及生物被膜细菌密度的分析(B)
图中不同字母表示在该初始细菌密度下,P. marina野生菌与ΔpilZ菌生物被膜的附着变态率或细菌密度组间有显著差异(p<0.05)
Fig. 2 Effect of wild-type and ΔpilZ biofilms formed with different initial bacterial density on larval settlement and metamorphosis (A) and the analysis of biofilm bacterial density (B)
Bars with different letters in the figure indicate that at the initial bacterial density, there’s a significant difference between the rate of larval settlement and metamorphosis or the bacterial density of P. marina wild-type and ΔpilZ biofilms (p<0.05)
表 1 本研究使用的细菌菌株和质粒
Tab. 1 The strains and plasmids used in this study
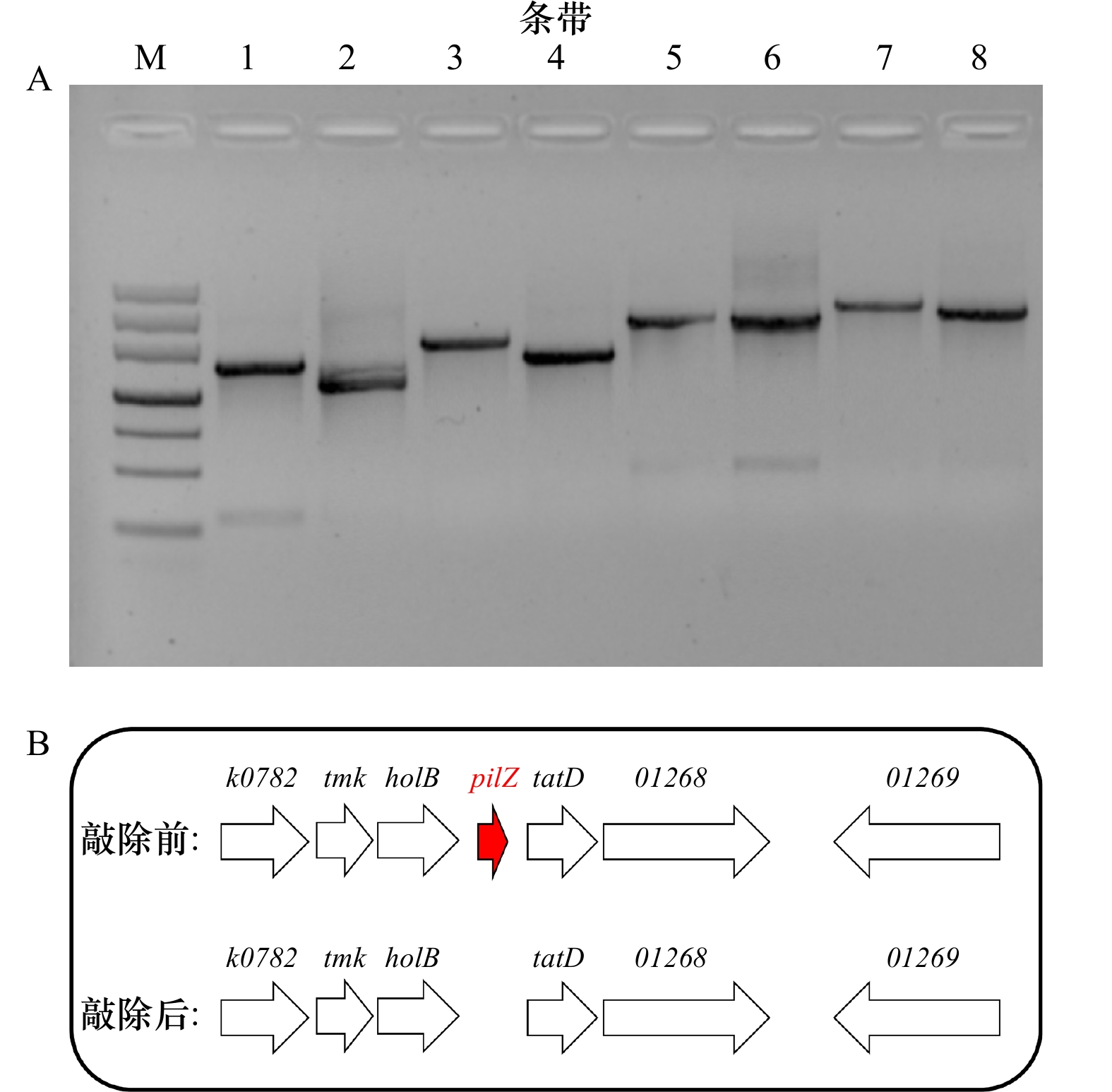
表 2 构建ΔpilZ菌株所使用的引物及其序列
Tab. 2 Primers used to construct ΔpilZ strain and its sequences
引物 序列(5'-3') pilZ-up-F CGGGATCCAGGTGAACTTGACCGAATA pilZ-up-R CGGAATTCGGTTAATCCTTTTATTTATT pilZ-down-F CGGAATTCTAAAAAACAGGCCCAATTTT pilZ-down-R AACTGCAGGACAATGCCTGAAATAGAAA pilZ-SF CCCTGTGGGTGTAGGTAA pilZ-SR CGTCGCGTGTATGAATAA pilZ-LF CGACCGTCACGACTTATC pilZ-LR TGTTCGCTGACACTTTGC 表 3 激光共聚焦显微镜荧光染料信息
Tab. 3 Laser confocal microscopy fluorescence dye information
荧光染料名称 结合物质 工作液浓度/
(μg·mL−1)波长范围/
nm碘化丙啶(PI) 死细菌 5 560~700 刀豆蛋白A(ConA-TMR) α-多糖 944.8 552~578 荧光增白剂(CFW M2R) β-多糖 189.0 254~432 DilC18(5)油(DiD`oil) 脂类 7.94 648~670 异硫氰酸荧光素异构体I(FITC) 蛋白质 46.6 495~519 -
[1] 李一峰, 杨金龙, 包卫洋, 等. 人工诱导物影响海洋无脊椎动物幼体附着变态的研究[J]. 海洋科学, 2011, 35(8): 102−107.Li Yifeng, Yang Jinlong, Bao Weiyang, et al. A review on artificial inducers influencing larval settlement and metamorphosis of marine invertebrates[J]. Marine Sciences, 2011, 35(8): 102−107. [2] Flemming H C, Wingender J. The biofilm matrix[J]. Nature Reviews Microbiology, 2010, 8(9): 623−633. doi: 10.1038/nrmicro2415 [3] Jenal U, Reinders A, Lori C. Cyclic di-GMP: second messenger extraordinaire[J]. Nature Reviews Microbiology, 2017, 15(5): 271−284. doi: 10.1038/nrmicro.2016.190 [4] Peng Lihua, Liang Xiao, Xu Jiakang, et al. Monospecific biofilms of Pseudoalteromonas promote larval settlement and metamorphosis of Mytilus coruscus[J]. Scientific Reports, 2020, 10(1): 2577. doi: 10.1038/s41598-020-59506-1 [5] Peng Lihua, Liang Xiao, Chang Ruiheng, et al. A bacterial polysaccharide biosynthesis-related gene inversely regulates larval settlement and metamorphosis of Mytilus coruscus[J]. Biofouling, 2020, 36(7): 753−765. doi: 10.1080/08927014.2020.1807520 [6] Liang Xiao, Zhang Xiukun, Peng Lihua, et al. The flagellar gene regulates biofilm formation and mussel larval settlement and metamorphosis[J]. International Journal of Molecular Sciences, 2020, 21(3): 710. doi: 10.3390/ijms21030710 [7] Hu Xiaomeng, Zhang Junbo, Ding Wenyang, et al. Reduction of mussel metamorphosis by inactivation of the bacterial thioesterase gene via alteration of the fatty acid composition[J]. Biofouling, 2021, 37(8): 911−921. doi: 10.1080/08927014.2021.1981882 [8] 蔡雨珊, 张秀坤, 竹攸汀, 等. 海假交替单胞菌(Pseudoalteromonas marina)鞭毛蛋白对生物被膜形成及厚壳贻贝附着的影响[J]. 海洋学报, 2021, 43(4): 75−83.Cai Yushan, Zhang Xiukun, Zhu Youting, et al. Effects of Pseudoalteromonas marina flagellin on biofilm formationand settlement of Mytilus coruscus[J]. Haiyang Xuebao, 2021, 43(4): 75−83. [9] Breaker R R. Prospects for riboswitch discovery and analysis[J]. Molecular Cell, 2011, 43(6): 867−879. doi: 10.1016/j.molcel.2011.08.024 [10] Matsuyama B Y, Krasteva P V, Baraquet C, et al. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa[J]. Proceedings of the National Academy of Sciences of the United States of America, 2016, 113(2): E209−E218. doi: 10.1073/pnas.1523148113 [11] Ryjenkov D A, Simm R, Römling U, et al. The PilZ domain is a receptor for the second messenger c-di-GMP: thePilZ domain protein YcgR controls motility in enterobacteria[J]. The Journal of Biological Chemistry, 2006, 281(41): 30310−30314. doi: 10.1074/jbc.C600179200 [12] Baker A E, Diepold A, Kuchma S L, et al. PilZ domain protein FlgZ mediates cyclic di-GMP-dependent swarming motility control in Pseudomonas aeruginosa[J]. Journal of Bacteriology, 2016, 198(13): 1837−1846. doi: 10.1128/JB.00196-16 [13] Valentini M, Filloux A. Multiple roles of c-di-GMP signaling in bacterial pathogenesis[J]. Annual Review of Microbiology, 2019, 73: 387−406. doi: 10.1146/annurev-micro-020518-115555 [14] Ross P, Weinhouse H, Aloni Y, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid[J]. Nature, 1987, 325(6101): 279−281. doi: 10.1038/325279a0 [15] Merighi M, Lee V T, Hyodo M, et al. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa[J]. Molecular Microbiology, 2007, 65(4): 876−895. doi: 10.1111/j.1365-2958.2007.05817.x [16] Klausen M, Heydorn A, Ragas P, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants[J]. Molecular Microbiology, 2003, 48(6): 1511−1524. doi: 10.1046/j.1365-2958.2003.03525.x [17] Guzzo C R, Salinas R K, Andrade M O, et al. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis[J]. Journal of Molecular Biology, 2009, 393(4): 848−866. doi: 10.1016/j.jmb.2009.07.065 [18] Peng Lihua, Liang Xiao, Guo Xingpan, et al. Complete genome of Pseudoalteromonas marina ECSMB14103, a mussel settlement-inducing bacterium isolated from the East China Sea[J]. Marine Genomics, 2018, 41: 46−49. doi: 10.1016/j.margen.2018.04.001 [19] Wang Pengxia, Yu Zichao, Li Baiyuan, et al. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas[J]. Microbial Cell Factories, 2015, 14: 11. doi: 10.1186/s12934-015-0194-8 [20] Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli[J]. Journal of Bacteriology, 1997, 179(2): 538−540. doi: 10.1128/jb.179.2.538-540.1997 [21] Zeng Zhenshun, Guo Xingpan, Li Baiyuan, et al. Characterization of self-generated variants in Pseudoalteromonas lipolytica biofilm with increased antifouling activities[J]. Applied Microbiology and Biotechnology, 2015, 99(23): 10127−10139. doi: 10.1007/s00253-015-6865-x [22] Yang Jinlong, Shen Peijing, Liang Xiao, et al. Larval settlement and metamorphosis of the mussel Mytilus coruscus in response to monospecific bacterial biofilms[J]. Biofouling, 2013, 29(3): 247−259. doi: 10.1080/08927014.2013.764412 [23] González-Machado C, Capita R, Riesco-Peláez F, et al. Visualization and quantification of the cellular and extracellular components of Salmonella agona biofilms at different stages of development[J]. PLoS One, 2018, 13(7): e0200011. doi: 10.1371/journal.pone.0200011 [24] Bobrov A G, Kirillina O, Ryjenkov D A, et al. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis[J]. Molecular Microbiology, 2011, 79(2): 533−551. doi: 10.1111/j.1365-2958.2010.07470.x [25] Hickman J W, Tifrea D F, Harwood C S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(40): 14422−14427. doi: 10.1073/pnas.0507170102 [26] Chua Songlin, Hultqvist L D, Yuan Mingjun, et al. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation[J]. Nature Protocols, 2015, 10(8): 1165−1180. doi: 10.1038/nprot.2015.067 [27] Whiteley C G, Lee D J. Bacterial diguanylate cyclases: structure, function and mechanism in exopolysaccharide biofilm development[J]. Biotechnology Advances, 2015, 33(1): 124−141. doi: 10.1016/j.biotechadv.2014.11.010 [28] Simm R, Morr M, Kader A, et al. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility[J]. Molecular Microbiology, 2004, 53(4): 1123−1134. doi: 10.1111/j.1365-2958.2004.04206.x [29] Hengge R, Gründling A, Jenal U, et al. Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers[J]. Journal of Bacteriology, 2016, 198(1): 15−26. doi: 10.1128/JB.00331-15 [30] Yoon S H, Waters C M. The ever-expanding world of bacterial cyclic oligonucleotide second messengers[J]. Current Opinion in Microbiology, 2021, 60: 96−103. doi: 10.1016/j.mib.2021.01.017 [31] Omadjela O, Narahari A, Strumillo J, et al. BcsA and BcsB form the catalytically active core of bacterial cellulose synthase sufficient for in vitro cellulose synthesis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(44): 17856−17861. doi: 10.1073/pnas.1314063110 [32] Richter A M, Possling A, Malysheva N, et al. Local c-di-GMP signaling in the control of synthesis of the E. coli biofilm exopolysaccharide pEtN-cellulose[J]. Journal of Molecular Biology, 2020, 432(16): 4576−4595. doi: 10.1016/j.jmb.2020.06.006 [33] Lee V T, Matewish J M, Kessler J L, et al. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production[J]. Molecular Microbiology, 2007, 65(6): 1474−1484. doi: 10.1111/j.1365-2958.2007.05879.x [34] Ghafoor A, Hay I D, Rehm B H A. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture[J]. Applied and Environmental Microbiology, 2011, 77(15): 5238−5246. doi: 10.1128/AEM.00637-11 [35] Ryder C, Byrd M, Wozniak D J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development[J]. Current Opinion in Microbiology, 2007, 10(6): 644−648. doi: 10.1016/j.mib.2007.09.010 [36] Liang Xiao, Zhang Junbo, Shao Anqi, et al. Bacterial cellulose synthesis gene regulates cellular c-di-GMP that control biofilm formation and mussel larval settlement[J]. International Biodeterioration & Biodegradation, 2021, 165: 105330. [37] Welker A, Cronenberg T, Zöllner R, et al. Molecular motors govern liquidlike ordering and fusion dynamics of bacterial colonies[J]. Physical Review Letters, 2018, 121(11): 118102. doi: 10.1103/PhysRevLett.121.118102 [38] Bonazzi D, Lo Schiavo V, Machata S, et al. Intermittent pili-mediated forces fluidize Neisseria meningitidis aggregates promoting vascular colonization[J]. Cell, 2018, 174(1): 143−155.e16. doi: 10.1016/j.cell.2018.04.010 [39] O’Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development[J]. Molecular Microbiology, 1998, 30(2): 295−304. doi: 10.1046/j.1365-2958.1998.01062.x [40] Semmler A B T, Whitchurch C B, Mattick J S. A re-examination of twitching motility in Pseudomonas aeruginosa[J]. Microbiology, 1999, 145(10): 2863−2873. doi: 10.1099/00221287-145-10-2863 [41] Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus[J]. Molecular Microbiology, 1997, 24(5): 885−893. doi: 10.1046/j.1365-2958.1997.4261783.x [42] Li Yinuo, Sun Hong, Ma Xiaoyuan, et al. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus[J]. Proceedings of the National Academy of Sciences of the United States of America, 2003, 100(9): 5443−5448. doi: 10.1073/pnas.0836639100 [43] Zeng Zhenshun, Guo Xingpan, Cai Xingsheng, et al. Pyomelanin from Pseudoalteromonas lipolytica reduces biofouling[J]. Microbial Biotechnology, 2017, 10(6): 1718−1731. doi: 10.1111/1751-7915.12773 -




 下载:
下载: