Isolation and identification of cultivable bacteria isolated from deep-sea sediment samples of the Indian Ocean
-
摘要: 为探索印度洋深海沉积物中纯培养细菌的多样性,本文对采自印度洋12个沉积物样品进行细菌纯培养分离,共获得343株细菌。所有细菌采用16S rRNA 基因进行比对分析,鉴定为4个门:厚壁菌门(Firmicutes)、变形菌门(Proteobacteria)、放线菌门(Actinobacteria)和拟杆菌门(Bacteroidetes),5个纲,13个目,26个科,39个属下的121个种。分离出的优势类群为链霉菌属(Streptomyces,分离率为16.53%)和芽孢杆菌属(Bacillus,分离率为8.26%),其中可能有12个潜在的新分类单元。研究结果表明,印度洋深海沉积环境中纯培养细菌资源丰富,潜在新物种较多;其次,分离培养方法与从海洋沉积物中获得的细菌物种多样性直接相关。Abstract: In order to explore the diversity of pure cultured bacteria in the deep-sea sedimentary environment of the Indian Ocean, bacteria were isolated and identified from 12 sediment samples collected from the Indian Ocean. A total of 343 strains of bacteria were obtained by pure culture isolation method and 16S rRNA gene sequence analysis showed that these isolates belonged to Firmicutes, Proteobacteria, Actinobacteria, Bacteroidete and were divided into 5 classes, 13 orders, 26 families and 121 species under 39 genera. The dominant taxa were Streptomyces (separation rate is 16.53%) and Bacillus (separation rate is 8.26%), among which there were 12 potential new taxa. The results show that the deep-sea sedimentary environment of the Indian Ocean is rich in culturable bacteria and contains many new species; secondly, the isolation and culture methods are directly related to the bacterial species diversity obtained from marine sediments.
-
Key words:
- Indian Ocean /
- sedimentary environment /
- bacteria /
- isolation and identification
-
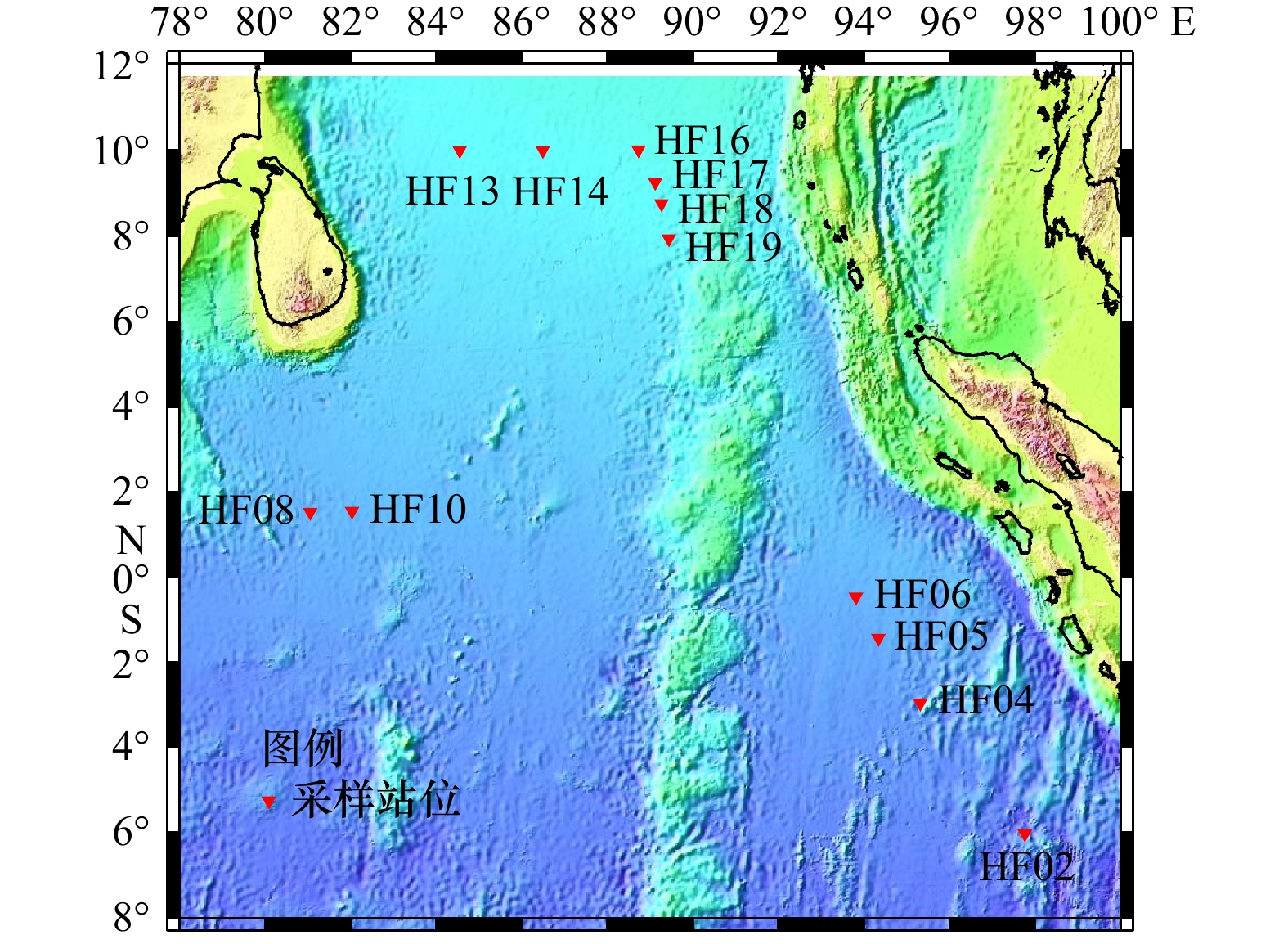
表 1 印度洋沉积物采样点信息
Tab. 1 Information of sampling sites from Indian Ocean sediment
站位名 纬度 经度 水深/m HF02 6°1′11.226″S 97°46′3.168″E 5 725 HF04 2°57′26.370″S 95°19′2.982″E 4 810 HF05 1°25′32.280″S 94°20′11.039″E 4 617 HF06 0°27′52.800″S 93°48′52.740″E 4 527 HF08 1°32′1.500″N 81°2′59.219″E 4 481 HF10 1°33′15.180″N 82°1′41.580″E 4 426 HF13 10°0′2.700″N 84°32′55.140″E 3 571 HF14 10°0′9.480″N 86°29′54.120″E 3 515 HF16 10°0′13.380″N 88°43′40.919″E 3 386 HF17 9°15′42.560″N 89°7′41.820″E 3 437 HF18 8°45′38.880″N 89°15′58.740″E 3 471 HF19 7°56′37.380″N 89°26′33.600″E 3 614 表 2 14种分离培养基
Tab. 2 14 kinds of culture media
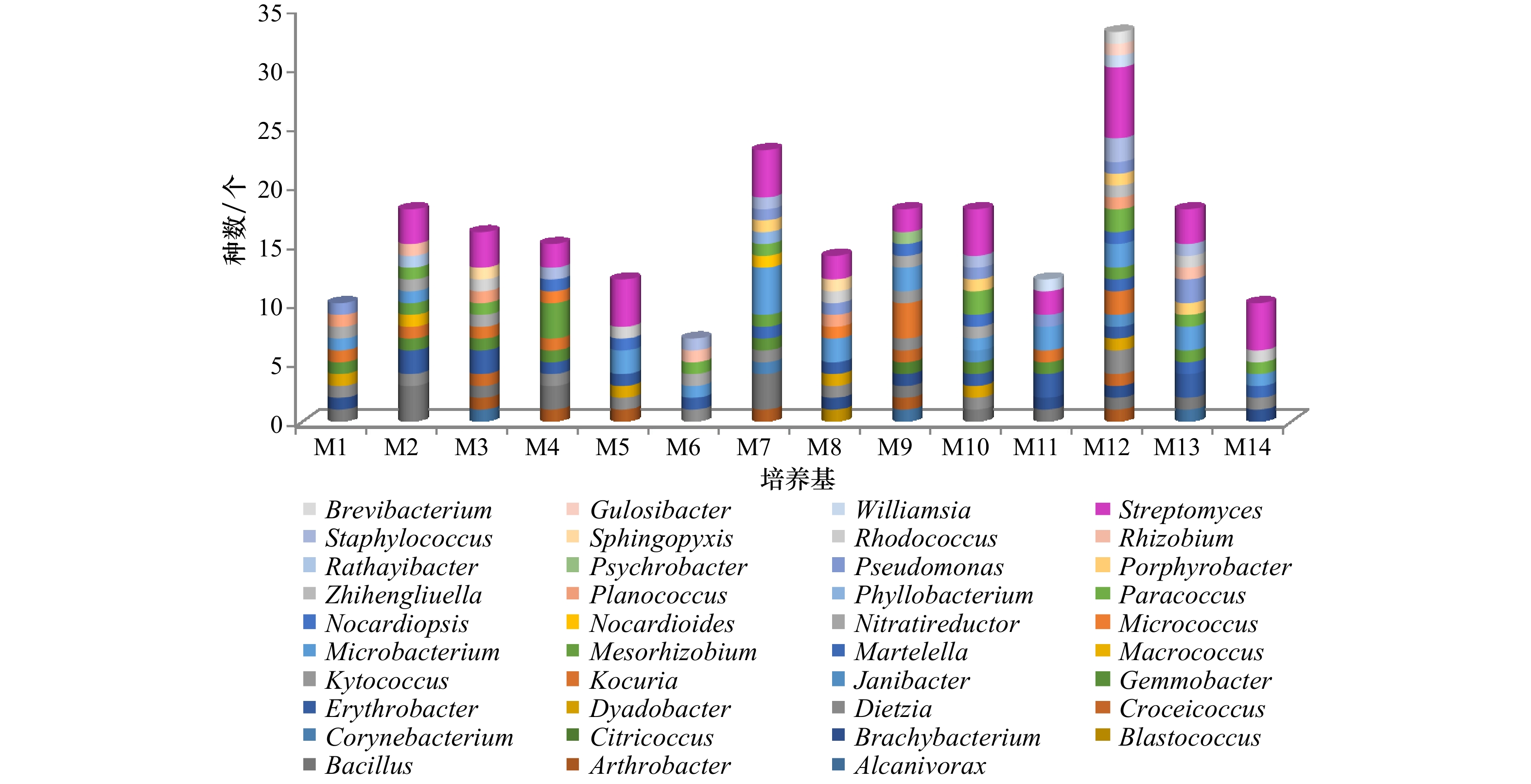
编号 名称 配方 M1 放线菌分离培养基 酪蛋白胨1 g、天冬酰胺0.1 g、丙酸钠4 g、K2HPO4 0.5 g、MgSO4·7H2O 0.1 g、 FeSO4·7H2O 1 mg、甘油1 mL、琼脂
15 g、双蒸水1 000 mL、pH 7.2~7.4M2 DA培养基 酪蛋白胨0.3 g、酵母膏0.1 g、葡萄糖0.01 g、琼脂15 g、双蒸水1 000 mL、pH 7.2~7.4 M3 高氏一号培养基 淀粉20 g、KNO3 1 g、K2HPO4 0.5 g、MgSO4·7H2O 0.5 g、NaCl 0.5 g、FeSO4 10 mg、琼脂15 g、双蒸水1 000 mL、pH 7.2~7.4 M4 淀粉−酪素培养基 淀粉10 g、酪蛋白0.3 g、KNO3 2 g、MgSO4·7H2O 0.05 g、K2HPO4 2 g、CaCO3 0.02 g、FeSO4 10 mg、琼脂 15 g、双蒸水1 000 mL、pH 7.2~7.4 M5 甘油−甘氨酸培养基 甘油20 g、甘氨酸2.5 g、K2HPO4 1 g、FeSO4 10 mg、MgSO4·7H2O 0.1 g、CaCO3 0.1 g、琼脂15 g、双蒸水1 000 mL、pH 7.4 M6 腐殖酸培养基 腐殖酸1.0 g、Na2HPO4 0.5 g、KCl 1.7 g、MgSO4·7H2O 0.05 g、FeSO4·7H2O 0.01 g、CaCl2 1 g、复合维生素 1 mL (维生素B1、维生素B2、烟酸、维生素B6、泛酸钙、肌糖、对氨基苯甲酸、维生素H 各0.5 mg、双蒸水1 000 mL)、琼脂15 g、双蒸水1 000 mL、pH 7.2 M7 甘油−精氨酸培养基 甘油5 g、精氨酸1 g、葡萄糖1 g、K2HPO4 0.3 g、MgSO4·7H2O 0.2 g、NaCl 0.3 g、复合维生素 1 mL (维生素B1、维生素B2、烟酸、维生素B6、泛酸钙、肌糖、对氨基苯甲酸、维生素H 各0.5 mg、双蒸水1 000 mL)、琼脂15 g、双蒸水1 000 mL、
pH 7.2M8 海藻糖−脯氨酸培养基 海藻糖5 g、脯氨酸1 g、(NH4)2SO4 1 g、CaCl2 2 g、K2HPO4 1 g、复合维生素1 mL (维生素B1、维生素B2、烟酸、维生素B6、泛酸钙、肌糖、对氨基苯甲酸、维生素H 各0.5 mg、双蒸水1 000 mL)、琼脂1g、双蒸水1 000 mL、pH 7.4 M9 牛肉膏−蛋白胨培养基 蛋白胨10 g、牛肉膏3 g、丙酮酸钠3 g、KCl 2 g、琼脂15 g、双蒸水1 000 mL、pH 7.0 M10 棉子糖−组氨酸培养基 棉子糖10 g、组氨酸1 g、MgSO4·7H2O 0.5 g、FeSO4·7H2O 0.01 g、琼脂15 g、双蒸水1 000 mL、pH 7.2 M11 改良海洋琼脂培养基 蛋白胨1 g、牛肉膏2 g、柠檬酸铁0.1 g、Na2CO3 0.16 g、柠檬酸钠4 mg、(NH4)NO3 1.6 mg、琼脂15 g、双蒸水1 000 mL、
pH 7.2M12 ISP2 培养基 牛肉膏0.4 g、葡萄糖0.4 g、麦芽膏0.4 g、复合维生素1 mL (维生素B1、维生素B2、烟酸、维生素B6、泛酸钙、肌糖、对氨基苯甲酸、维生素H 各0.5 mg、双蒸水1 000 mL )、琼脂15 g、双蒸水1 000 mL、pH 8.0 M13 R2A培养基 牛肉膏0.5 g、眎蛋白胨0.5 g、酪氨酸0.5 g、葡萄糖0.5 g、可溶性淀粉0.5 g、丙酮酸钠0.3 g、K2HPO4 0.3 g、MgSO4·7H2O
0.05 g、琼脂15 g、双蒸水1 000 mL、pH 7.2M14 海盐琼脂培养基 自来水1 000 mL、琼脂15 g、pH 7.2 表 3 分离菌株整体分布表
Tab. 3 Overall distribution of isolated strains
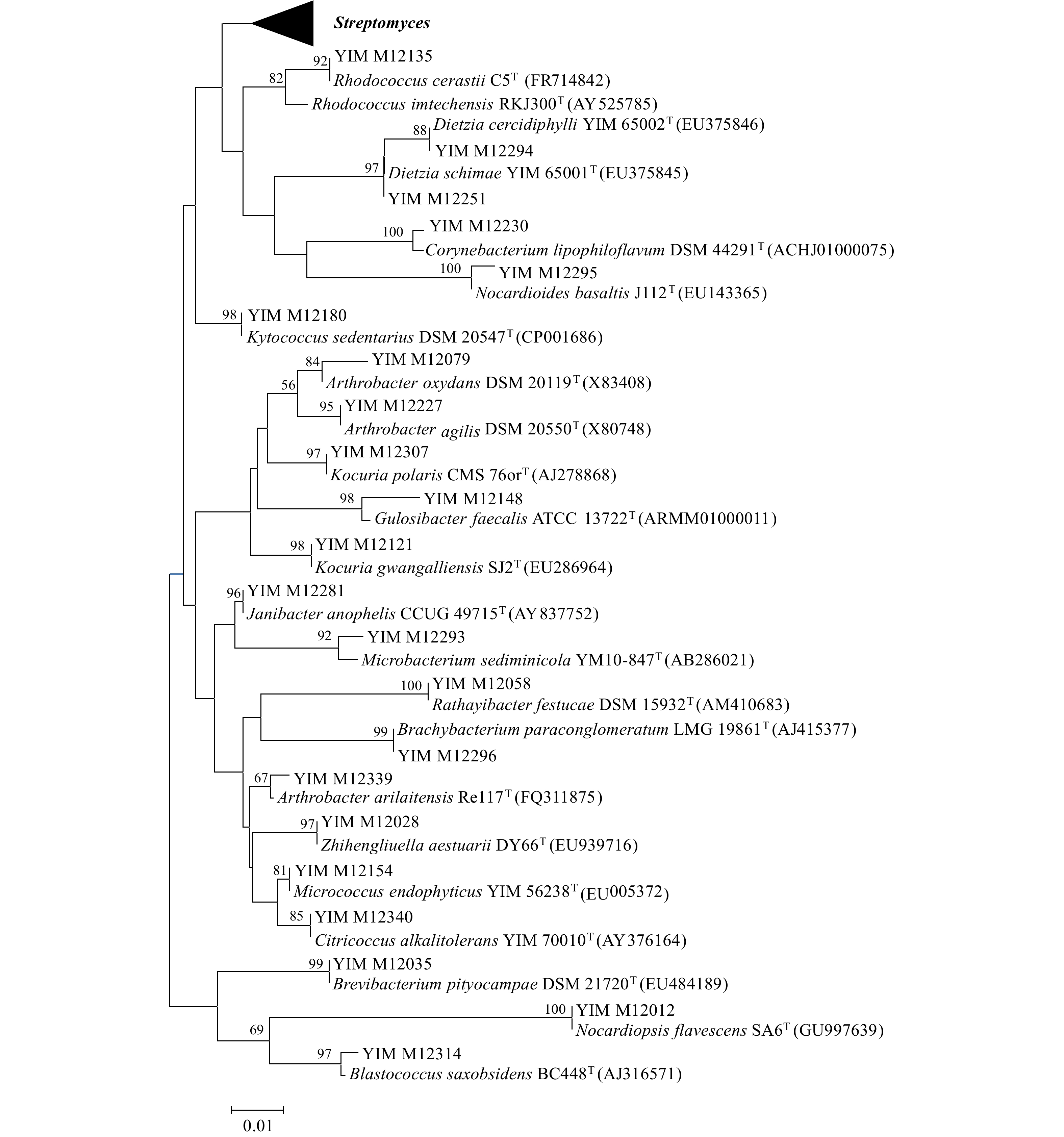
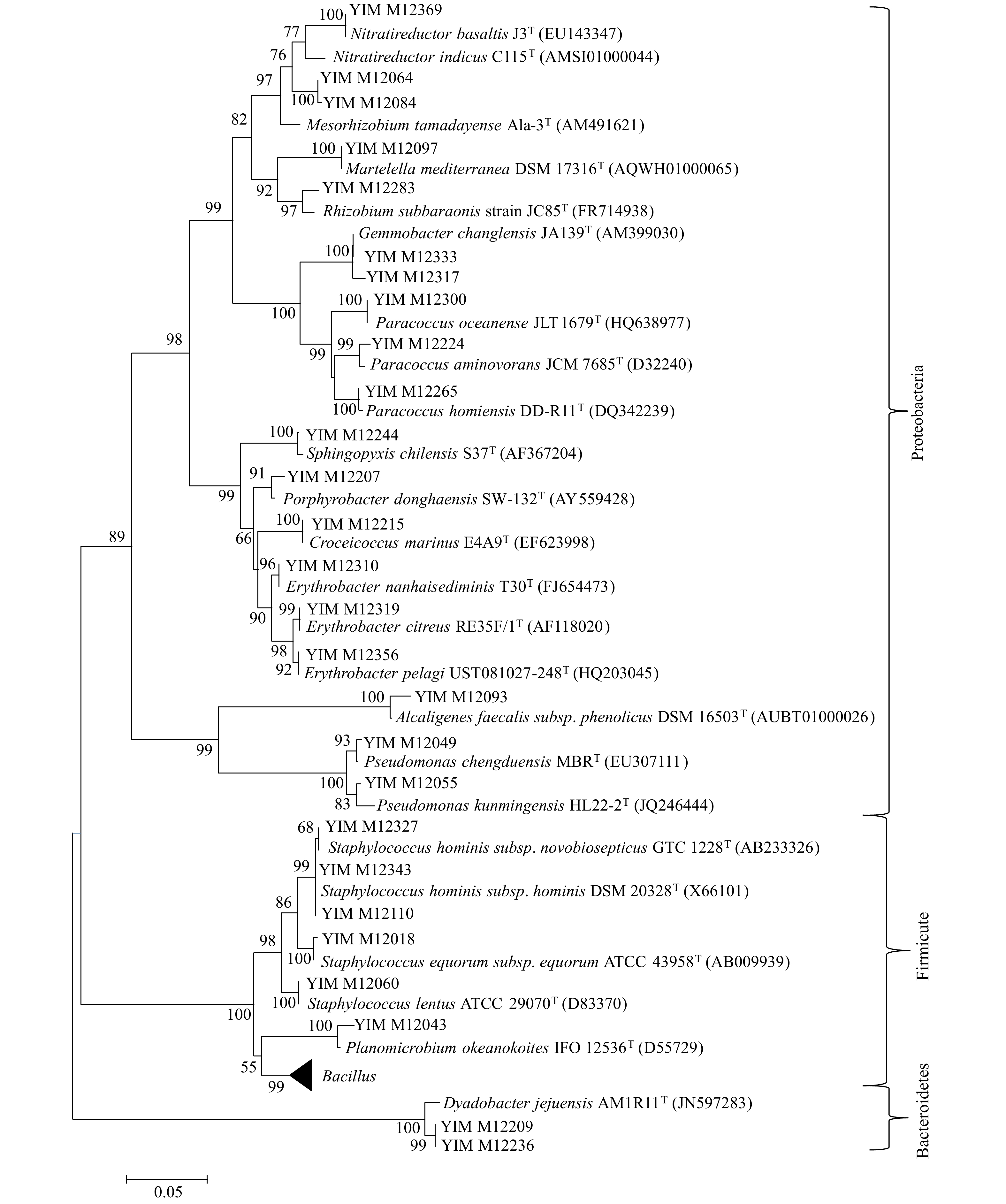
门 纲 目 科 属 种 占总种数百分比/% 放线菌门 1 6 13 20 71 58.7 厚壁菌门 1 1 3 4 21 17.4 拟杆菌门 1 1 1 1 3 2.4 变形菌门 2 5 9 14 26 21.5 总计 5 13 26 39 121 100 表 4 种分布百分比
Tab. 4 Percentages of species distribution
属名 种数 百分比/% 属名 种数 百分比/% Streptomyces 20 16.53 Gemmobacter 2 1.65 Bacillus 10 8.26 Corynebacterium 1 0.83 Kocuria 6 4.96 Williamsia 1 0.83 Staphylococcus 6 4.96 Brevibacterium 1 0.83 Dietzia 5 4.13 Brachybacterium 1 0.83 Rhodococcus 5 4.13 Kytococcus 1 0.83 Microbacterium 5 4.13 Rathayibacter 1 0.83 Arthrobacter 5 4.13 Citricoccus 1 0.83 Nocardioides 5 4.13 Zhihengliuella 1 0.83 Nocardiopsis 5 4.13 Blastococcus 1 0.83 Planococcus 4 3.31 Macrococcus 1 0.83 Paracoccus 4 3.31 Croceicoccus 1 0.83 Pseudomonas 4 3.31 Porphyrobacter 1 0.83 Dyadobacter 3 2.48 Sphingopyxis 1 0.83 Erythrobacter 3 2.48 Martelella 1 0.83 Mesorhizobium 3 2.48 Nitratireductor 1 0.83 Janibacter 2 1.65 Phyllobacterium 1 0.83 Gulosibacter 2 1.65 Psychrobacter 1 0.83 Micrococcus 2 1.65 Alcanivorax 1 0.83 Rhizobium 2 1.65 表 5 潜在新物种的16S rRNA基因序列比对结果
Tab. 5 Comparison of 16S rRNA gene sequences of potential new species
菌号 相似菌株 相似性/% YIM M12043 Planococcus donghaensis DSM 22276T 97.27 YIM M12055 Pseudomonas kunmingensis HL22-2T 95.58 YIM M12064 Mesorhizobium thiogangeticum SJTT 97.37 YIM M12082 Dyadobacter tibetensis CGMCC 1.12215T 97.99 YIM M12084 Mesorhizobium metallidurans STM 2683T 97.05 YIM M12100 Mesorhizobium thiogangeticum DSM 17097T 97.28 YIM M12113 Arthrobacter oxydans ATCC 14358T 97.87 YIM M12116 Phyllobacterium myrsinacearum ATCC 43590T 97.28 YIM M12122 Gemmobacter changlensis CCUG 53722T 94.88 YIM M12139 Alcanivorax dieselolei CGMCC 1.3690T 92.66 YIM M12140 Macrococcus brunensis CCM 4811T 91.40 YIM M12148 Gulosibacter faecalis ATCC 13722T 97.05 -
[1] Arrigo K R. Marine microorganisms and global nutrient cycles[J]. Nature, 2005, 437(7057): 349−355. doi: 10.1038/nature04159 [2] Bowler C, Karl D M, Colwell R R. Microbial oceanography in a sea of opportunity[J]. Nature, 2009, 459(7244): 180−184. doi: 10.1038/nature08056 [3] 张偲, 等. 中国海洋微生物多样性[M]. 1版. 北京: 科学出版社, 2013.Zhang Si, et al. Diversities of Marine Microbes in China[M]. 1st ed. Beijing: Science Press, 2013. [4] Rochelle P A, Cragg B A, Fry J C, et al. Effect of sample handling on estimation of bacterial diversity in marine sediments by 16S rRNA gene sequence analysis[J]. FEMS Microbiology Ecology, 1994, 15(1/2): 215−225. [5] Wellsbury P, Mather I, Parkes R J. Geomicrobiology of deep, low organic carbon sediments in the Woodlark Basin, Pacific Ocean[J]. FEMS Microbiology Ecology, 2002, 42(1): 59−70. doi: 10.1111/j.1574-6941.2002.tb00995.x [6] Newberry C J, Webster G, Cragg B A, et al. Diversity of prokaryotes and Methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190[J]. Environmental Microbiology, 2004, 6(3): 274−287. doi: 10.1111/j.1462-2920.2004.00568.x [7] Inagaki F, Nunoura T, Nakagawa S, et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean margin[J]. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(8): 2815−2820. doi: 10.1073/pnas.0511033103 [8] Bidle K A, Kastner M, Bartlett D H. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia margin (ODP site 892B)[J]. FEMS Microbiology Letters, 1999, 177(1): 101−108. doi: 10.1111/j.1574-6968.1999.tb13719.x [9] Inagaki F, Suzuki M, Takai K, et al. Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of Okhotsk[J]. Applied and Environmental Microbiology, 2003, 69(12): 7224−7235. doi: 10.1128/AEM.69.12.7224-7235.2003 [10] Heijs S K, Haese R R, Van Der Wielen P W J J, et al. Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a Mediterranean cold seep[J]. Microbial Ecology, 2007, 53(3): 384−398. doi: 10.1007/s00248-006-9172-3 [11] 徐丽华, 娄恺, 张华, 等. 微生物资源学[M]. 2版. 北京: 科学出版社, 2010: 90.Xu Lihua, Lou Kai, Zhang Hua, et al. Microbial Resources[M]. 2nd ed. Beijing: Science Press, 2010: 90. [12] Arrieta J M, Arnaud-haond S, Duarte C M. What lies underneath: Conserving the oceans’ genetic resources[J]. Proceedings of the National Academy of Sciences of the United States of America, 2010, 107(43): 18318−18324. doi: 10.1073/pnas.0911897107 [13] Schut F, De Vries E J, Gottschal J C, et al. Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions[J]. Applied Environmental Microbiology, 1993, 59(7): 150−161. [14] 王宏梅, 赵心清. 可培养海洋放线菌生物多样性的研究进展[J]. 微生物学通报, 2007, 34(5): 996−1000. doi: 10.3969/j.issn.0253-2654.2007.05.040Wang Hongmei, Zhao Xinqing. Progress in the bio-diversity studies of culturable marine actinobacteria[J]. Microbiology China, 2007, 34(5): 996−1000. doi: 10.3969/j.issn.0253-2654.2007.05.040 [15] Mincer T J, Jensen P R, Kauffman C A, et al. Widespread and persistent populations of a major new marine actinomycete taxon in ocean sediments[J]. Applied and Environmental Microbiology, 2002, 68(10): 5005−5011. doi: 10.1128/AEM.68.10.5005-5011.2002 [16] Terahara T, Kobayashi T, Imada C. An effective method based on wet-heat treatment for the selective isolation of micromonospora from estuarine sediments[J]. World Journal of Microbiology and Biotechnology, 2013, 29(9): 1677−1684. doi: 10.1007/s11274-013-1330-4 [17] Li Wenjun, Xu Ping, Schumann P, et al. Georgenia ruanii sp. nov. , a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia[J]. International Journal of Systematic and Evolutionary Microbiology, 2007, 57(Pt7): 1424−1428. [18] Messick J B, Berent L M, Cooper S K. Development and evaluation of a PCR-based assay for detection of Haemobartonella felis in cats and differentiation of H. felis from related bacteria by restriction fragment length polymorphism analysis[J]. Journal of Clinical Microbiology, 1998, 36(2): 462−466. doi: 10.1128/JCM.36.2.462-466.1998 [19] Kim O S, Cho Y J, Lee K, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species[J]. International Journal of Systematic and Evolutionary Microbiology, 2012, 62(3): 716−721. [20] Thompson J D, Gibson T J, Plewniak F, et al. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Research, 1997, 25(24): 4876−4882. doi: 10.1093/nar/25.24.4876 [21] Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Molecular Biology and Evolution, 2016, 33(7): 1870−1874. doi: 10.1093/molbev/msw054 [22] Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Molecular Biology and Evolution, 1987, 4(4): 406−425. [23] Wayne L G, Brenner D J, Colwell R R, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics[J]. International Journal of Systematic Bacteriology, 1987, 37(4): 463−464. [24] Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards[J]. Microbiology Today, 2006, 8(4): 6−9. [25] Goodfellow M, Fiedler H P. A guide to successful bioprospecting: informed by actinobacterial systematics[J]. Antonie van Leeuwenhoek, 2010, 98(2): 119−142. doi: 10.1007/s10482-010-9460-2 [26] 徐盈. 南海沉积环境细菌分离方法及多样性研究[D]. 昆明: 云南大学, 2012.Xu Ying. Study on isolation methods and bacterial diversity in sediment environments from South China Sea[D]. Kunming: Yunnan University, 2012. [27] 张偲, 张长生, 田新朋, 等. 中国海洋微生物多样性研究[J]. 中国科学院院刊, 2010, 25(6): 651−658. doi: 10.3969/j.issn.1000-3045.2010.06.011Zhang Si, Zhang Changsheng, Tian Xinpeng, et al. The study of diversities of marine microbes in China[J]. Bulletin of the Chinese Academy of Sciences, 2010, 25(6): 651−658. doi: 10.3969/j.issn.1000-3045.2010.06.011 -




 下载:
下载: