Isolation and diversity analysis of coral-associated DMSP degrading bacteria
-
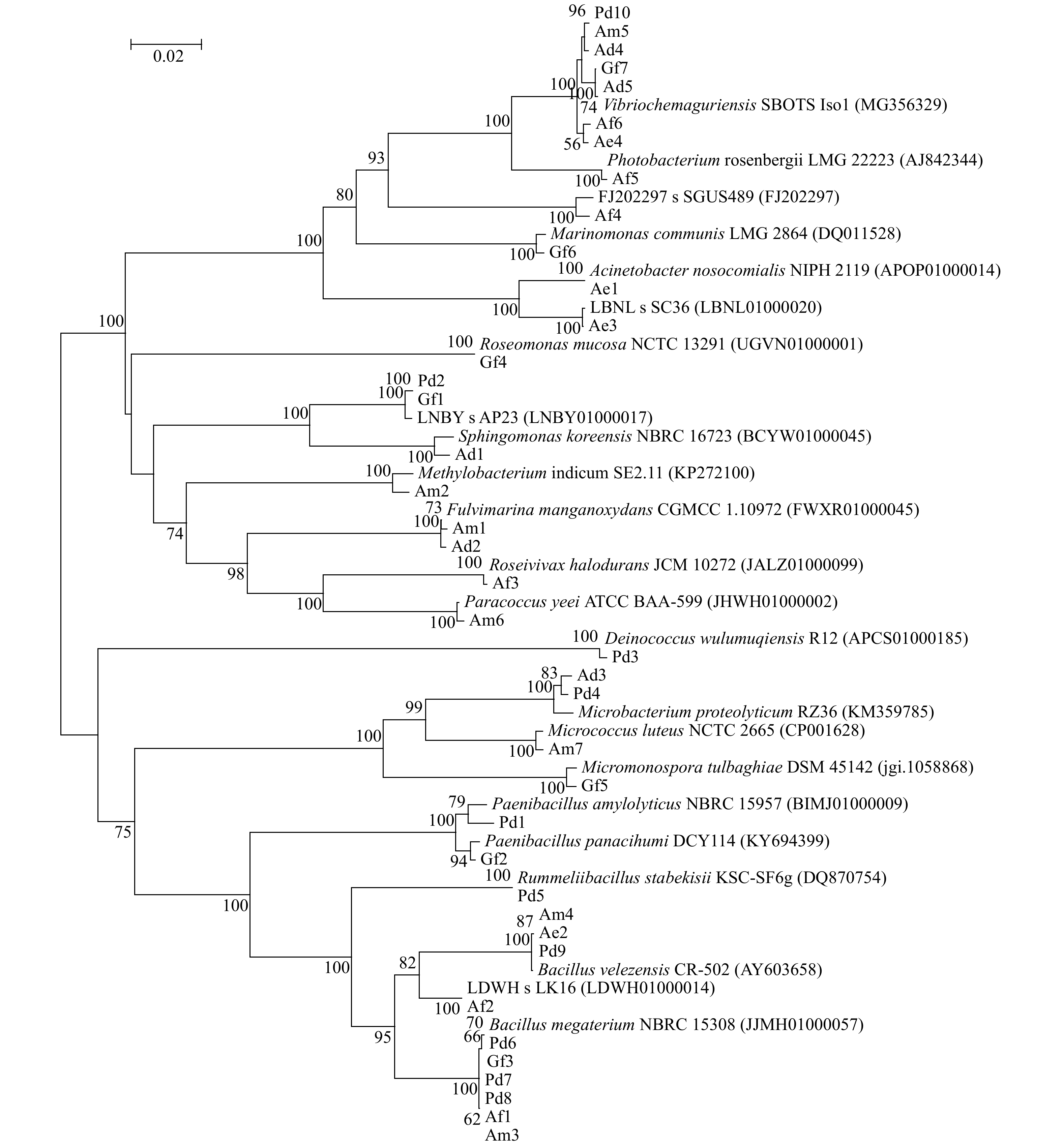
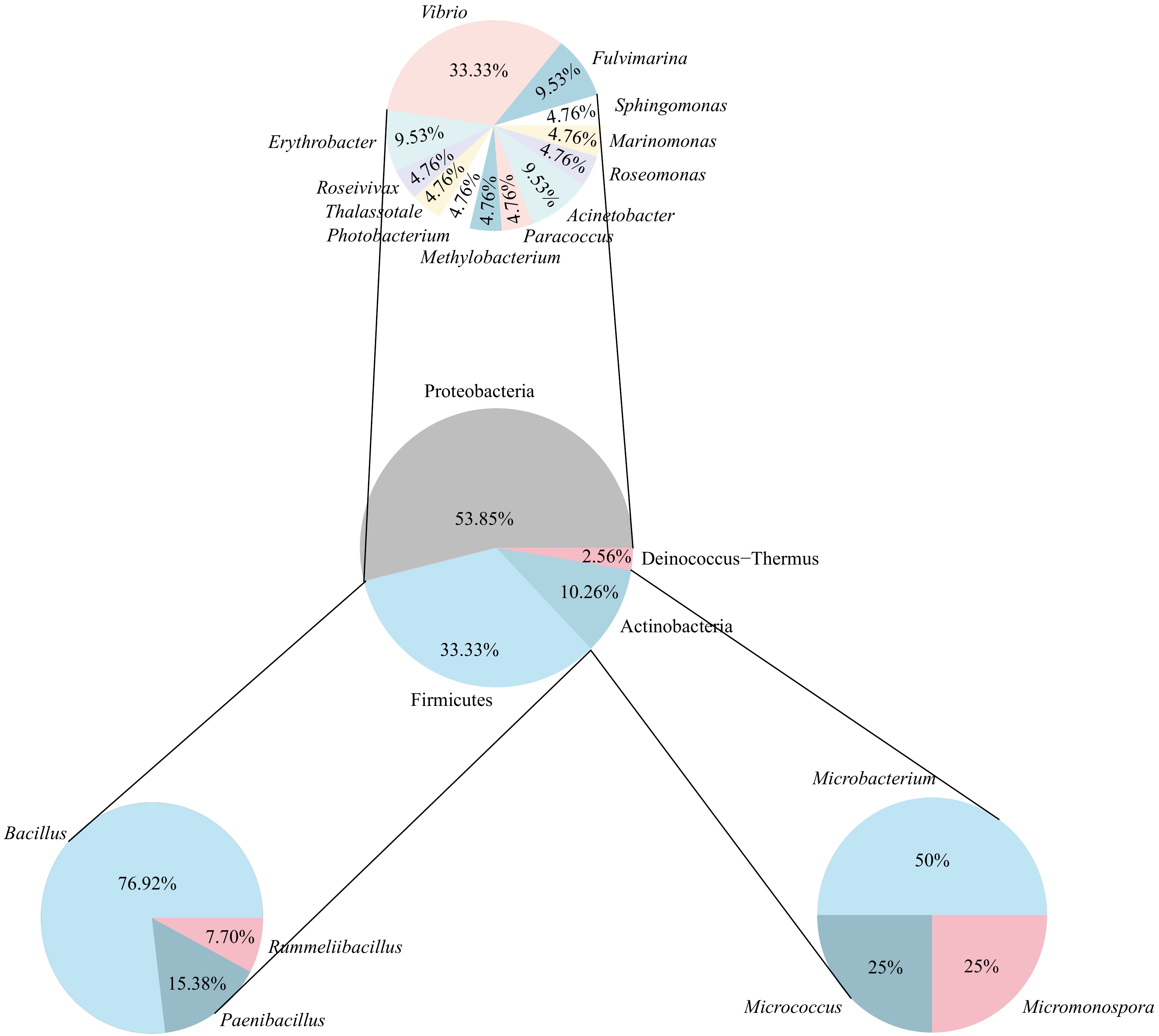
摘要: 二甲基巯基丙酸内盐(DMSP)是地球上最丰富的有机硫分子之一,在全球硫循环和气候调节中具有重要的作用。DMSP是“冷室气体”二甲基硫(DMS)最主要的前体物质;在海洋中,DMSP可被多种途径降解,微生物降解是其最重要的途径之一。珊瑚礁是海洋DMS重要的来源之一,珊瑚共附生DMSP降解菌在DMS生产过程中发挥着重要的作用。本研究从多孔鹿角珊瑚(Acropora millepora)、美丽鹿角珊瑚(Acropora formosa)、多棘鹿角珊瑚(Acropora echinata)、指状鹿角珊瑚(Acropora digitifera)、鹿角杯形珊瑚(Pocillopora damicornis)和丛生盔形珊瑚(Galaxea fascicularis) 6种造礁石珊瑚中分离获得珊瑚共附生DMSP降解菌39株,基于16S rRNA基因序列对DMSP降解菌进行系统发育分析,39株DMSP降解菌株分别隶属于4个门、6个纲、19个属,优势属为芽孢杆菌属(Bacillus);通过火焰光度检测器−气相色谱(GC-FPD)联用技术检测DMSP降解产物,分析DMSP降解菌的DMS生产能力,结果显示,9株菌具有高产DMS能力,高产DMS菌株对于珊瑚应对气候变暖的益生作用有待后续深入研究。Abstract: Dimethylsulfoniopropionate (DMSP) is one of the most abundant organic sulfur molecules on earth, and plays an important role in global sulfur cycle and climate regulation. DMSP is the principal precursor of dimethyl sulfide (DMS), which can be degraded through many ways in the ocean, and microbial degradation is one of the most important ways. Coral reef is one of the major sources of marine DMS, and coral-associated DMSP degrading bacteria play an important role in the process of DMS production. In the present study, 39 strains of DMSP degrading bacteria were isolated from six reef building corals, including Acropora millepora, Acropora formosa, Acropora echinata, Acropora digitifera, Pocillopora damicornis, and Galaxea fascicularis. The phylogenetic analysis of DMSP degrading bacteria was performed based on 16S rRNA gene sequence, 39 strains of DMSP degrading bacteria belonged to 4 phyla, 6 classes, and 19 genera, the dominant genus was Bacillus. The DMS production efficiency of DMSP degrading bacteria were analyzed by GC-FPD detection of DMSP products, the results showed that 9 strains had the ability of high DMS production. The probiotic effects of bacteria with high DMS production on corals in response to climate warming need to be further studied.
-
Key words:
- DMSP /
- DMS /
- degrading bacteria /
- coral reef ecosystems /
- sulfur cycle
-
表 1 6种石珊瑚中分离获得菌株在属水平上的分布
Tab. 1 The distribution of strains isolated from six species of corals at genus level
属 鹿角杯形珊瑚 丛生盔形珊瑚 多孔鹿角珊瑚 指状鹿角珊瑚 美丽鹿角珊瑚 多棘鹿角珊瑚 Acinetobacter − − − − − 2 Bacillus 4 1 2 − 2 1 Deinococcus 1 − − − − − Erythrobacter 1 1 − − − − Fulvimarina − − 1 1 − − Marinomonas − 1 − − − − Methylobacterium − − 1 − − − Micrococcus − − 1 − − − Micromonospora − 1 − − − − Microbacterium 1 − − 1 − − Paenibacillus 1 1 − − − − Photobacterium − − − − 1 − Paracoccus − − 1 − − − Roseomonas − 1 − − − − Roseivivax − − − − 1 − Rummeliibacillus 1 − − − − − Sphingomonas − − − 1 − − Thalassotale − − − − 1 − Vibrio 1 1 1 2 1 1 注:−表示没有获得隶属于该属的菌株。 -
[1] Hoegh-Guldberg O, Mumby P J, Hooten A J, et al. Coral reefs under rapid climate change and ocean acidification[J]. Science, 2007, 318(5857): 1737−1742. doi: 10.1126/science.1152509 [2] Hughes T P, Baird A H, Bellwood D R, et al. Climate change, human impacts, and the resilience of coral reefs[J]. Science, 2003, 301(5635): 929−933. doi: 10.1126/science.1085046 [3] Ateweberhan M, Mcclanahan T R, Graham N A J, et al. Episodic heterogeneous decline and recovery of coral cover in the Indian Ocean[J]. Coral Reefs, 2011, 30(3): 739−752. doi: 10.1007/s00338-011-0775-x [4] Panithanarak T. Effects of the 2010 coral bleaching on phylogenetic clades and diversity of zooxanthellae (Symbiodinium spp.) in soft corals of the genus Sinularia[J]. Plankton and Benthos Research, 2015, 10(1): 11−17. doi: 10.3800/pbr.10.11 [5] Teixeira C D, Leitão R L L, Ribeiro F V, et al. Sustained mass coral bleaching (2016−2017) in Brazilian turbid-zone reefs: taxonomic, cross-shelf and habitat-related trends[J]. Coral Reefs, 2019, 38(4): 801−813. doi: 10.1007/s00338-019-01789-6 [6] Zhang Xiaohua, Liu Ji, Liu Jingli, et al. Biogenic production of DMSP and its degradation to DMS—their roles in the global sulfur cycle[J]. Science China Life Sciences, 2019, 62(10): 1296−1319. doi: 10.1007/s11427-018-9524-y [7] Charlson R J, Lovelock J E, Andreae M O, et al. Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate[J]. Nature, 1987, 326(6114): 655−661. doi: 10.1038/326655a0 [8] Weis V M. Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis[J]. Journal of Experimental Biology, 2008, 211(19): 3059−3066. doi: 10.1242/jeb.009597 [9] Sunda W, Kieber D J, Kiene R P, et al. An antioxidant function for DMSP and DMS in marine algae[J]. Nature, 2002, 418(6895): 317−320. doi: 10.1038/nature00851 [10] Raina J B, Tapiolas D M, Forêt S, et al. DMSP biosynthesis by an animal and its role in coral thermal stress response[J]. Nature, 2013, 502(7473): 677−680. doi: 10.1038/nature12677 [11] Curson A R J, Todd J D, Sullivan M J, et al. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes[J]. Nature Reviews Microbiology, 2011, 9(12): 849−859. doi: 10.1038/nrmicro2653 [12] Liu Jingli, Liu Ji, Zhang Shenghui, et al. Novel insights into bacterial dimethylsulfoniopropionate catabolism in the East China Sea[J]. Frontiers in Microbiology, 2018, 9: 3206. doi: 10.3389/fmicb.2018.03206 [13] Howard E C, Henriksen J R, Buchan A, et al. Bacterial taxa that limit sulfur flux from the ocean[J]. Science, 2006, 314(5799): 649−652. doi: 10.1126/science.1130657 [14] Curson A R J, Rogers R, Todd J D, et al. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate-changing gas dimethylsulfide in several marine α-proteobacteria and Rhodobacter sphaeroides[J]. Environmental Microbiology, 2008, 10(3): 757−767. doi: 10.1111/j.1462-2920.2007.01499.x [15] Reisch C R, Moran M A, Whitman W B. Bacterial catabolism of dimethylsulfoniopropionate (DMSP)[J]. Frontiers in Microbiology, 2011, 2: 172. [16] Todd J D, Rogers R, Li Youguo, et al. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria[J]. Science, 2007, 315(5812): 666−669. doi: 10.1126/science.1135370 [17] Raina J B, Tapiolas D, Willis B L, et al. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur[J]. Applied and Environmental Microbiology, 2009, 75(11): 3492−3501. doi: 10.1128/AEM.02567-08 [18] Tandon K, Lu C Y, Chiang P W, et al. Comparative genomics: dominant coral-bacterium Endozoicomonas acroporae metabolizes dimethylsulfoniopropionate (DMSP)[J]. The ISME Journal, 2020, 14(5): 1290−1303. doi: 10.1038/s41396-020-0610-x [19] Garren M, Son K, Raina J B, et al. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals[J]. The ISME Journal, 2014, 8(5): 999−1007. doi: 10.1038/ismej.2013.210 [20] Zhao Meixia, Yu Kefu, Zhang Qiaomin, et al. Long-term decline of a fringing coral reef in the northern South China Sea[J]. Journal of Coastal Research, 2012, 28(5): 1088−1099. doi: 10.2112/JCOASTRES-D-10-00172.1 [21] Yuan Xiangcheng, Guo Yajuan, Cai Weijun, et al. Coral responses to ocean warming and acidification: implications for future distribution of coral reefs in the South China Sea[J]. Marine Pollution Bulletin, 2019, 138: 241−248. doi: 10.1016/j.marpolbul.2018.11.053 [22] Souza M P D, Yoch D C. Purification and characterization of dimethylsulfoniopropionate lyase from an alcaligenes-like dimethyl sulfide-producing marine isolate[J]. Applied and Environmental Microbiology, 1995, 61(1): 21−26. doi: 10.1128/AEM.61.1.21-26.1995 [23] Todd J D, Curson A R J, Dupont C L, et al. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi[J]. Environmental Microbiology, 2009, 11(6): 1624−1625. doi: 10.1111/j.1462-2920.2009.01919.x [24] Zhang Shenghui, Yu Juan, Ding Qiongyao, et al. Effect of elevated pCO2 on trace gas production during an ocean acidification mesocosm experiment[J]. Biogeosciences, 2018, 15(21): 6649−6658. doi: 10.5194/bg-15-6649-2018 [25] Yang Guipeng, Zhang Shenghui, Zhang Honghai, et al. Distribution of biogenic sulfur in the Bohai Sea and northern Yellow Sea and its contribution to atmospheric sulfate aerosol in the late fall[J]. Marine Chemistry, 2015, 169: 23−32. doi: 10.1016/j.marchem.2014.12.008 [26] Bosch T C G, Mcfall-Ngai M J. Metaorganisms as the new frontier[J]. Zoology, 2011, 114(4): 185−190. doi: 10.1016/j.zool.2011.04.001 [27] Benavides M, Bednarz V N, Ferrier-Pagès C. Diazotrophs: overlooked key players within the coral symbiosis and tropical reef ecosystems?[J]. Frontiers in Marine Science, 2017, 4: 10. [28] Rypien K L, Ward J R, Azam F. Antagonistic interactions among coral-associated bacteria[J]. Environmental Microbiology, 2010, 12(1): 28−39. doi: 10.1111/j.1462-2920.2009.02027.x [29] Nissimov J, Rosenberg E, Munn C B. Antimicrobial properties of resident coral mucus bacteria of Oculina patagonica[J]. FEMS Microbiology Letters, 2009, 292(2): 210−215. doi: 10.1111/j.1574-6968.2009.01490.x [30] Tang Kaihao, Wang Yan, Wang Xiaoxue. Recent progress on signalling molecules of coral-associated microorganisms[J]. Science China Earth Science, 2019, 62(4): 609−618. doi: 10.1007/s11430-018-9332-1 [31] Bourne D G, Dennis P G, Uthicke S, et al. Coral reef invertebrate microbiomes correlate with the presence of photosymbionts[J]. The ISME Journal, 2013, 7(7): 1452−1458. doi: 10.1038/ismej.2012.172 [32] González J M, Kiene R P, Moran M A. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the α-subclass of the class Proteobacteria[J]. Applied and Environmental Microbiology, 1999, 65(9): 3810−3819. doi: 10.1128/AEM.65.9.3810-3819.1999 [33] Hernandez-Agreda A, Gates R D, Ainsworth T D. Defining the core microbiome in corals' microbial soup[J]. Trends in Microbiology, 2017, 25(2): 125−140. doi: 10.1016/j.tim.2016.11.003 [34] Rosenberg E, Koren O, Reshef L, et al. The role of microorganisms in coral health, disease and evolution[J]. Nature Reviews Microbiology, 2007, 5(5): 355−362. doi: 10.1038/nrmicro1635 [35] Wagner C, Stadtman E R. Bacterial fermentation of dimethyl-β-propiothetin[J]. Archives of Biochemistry & Biophysics, 1962, 98(2): 331−336. [36] 张斯颖, 陈宇丰, 骆焱平, 等. 丛生盔形珊瑚共附生放线菌NLG-3的抑菌活性及鉴定[J]. 西北农林科技大学学报(自然科学版), 2019, 47(10): 97−105.Zhang Siying, Chen Yufeng, Luo Yanping, et al. Antibacterial activity and identification of epiphytic actinomycete NLG-3 from Galaxea facicularis[J]. Journal of Northwest A & F University (Natural Science Edition), 2019, 47(10): 97−105. [37] 林钰, 刘荣华, 周顺, 等. 马里亚纳海沟沉积物可培养异养细菌的多样性及其DMSP降解能力[J]. 微生物学报, 2021, 61(4): 828−844.Lin Yu, Liu Ronghua, Zhou Shun, et al. Diversity of culturable heterotrophic bacteria from sediments of the Mariana Trench and their ability to degrade dimethylsulfoniopropionate (DMSP)[J]. Acta Microbiologica Sinica, 2021, 61(4): 828−844. [38] Kooperman N, Ben-Dov E, Kramarsky-Winter E, et al. Coral mucus-associated bacterial communities from natural and aquarium environments[J]. FEMS Microbiology Letters, 2007, 276(1): 106−113. doi: 10.1111/j.1574-6968.2007.00921.x [39] Zeng Yinxin, Qiao Zongyun, Yu Yong, et al. Diversity of bacterial dimethylsulfoniopropionate degradation genes in surface seawater of Arctic Kongsfjorden[J]. Scientific Reports, 2016, 6: 33031. doi: 10.1038/srep33031 [40] Schnicker N J, De Silva S M, Todd J D, et al. Structural and biochemical insights into dimethylsulfoniopropionate cleavage by cofactor-bound DddK from the prolific marine bacterium Pelagibacter[J]. Biochemistry, 2017, 56(23): 2873−2885. doi: 10.1021/acs.biochem.7b00099 [41] Newton R J, Griffin L E, Bowles K M, et al. Genome characteristics of a generalist marine bacterial lineage[J]. The ISME Journal, 2010, 4(6): 784−798. doi: 10.1038/ismej.2009.150 [42] Miller T R, Belas R. Dimethylsulfoniopropionate metabolism by Pfiesteria-associated Roseobacter spp.[J]. Applied and Environmental Microbiology, 2004, 70(6): 3383−3391. doi: 10.1128/AEM.70.6.3383-3391.2004 [43] 孙浩, 谭斯尹, 梁金昌, 等. 中国东海二甲基巯基丙酸内盐(DMSP)合成与降解菌的水平和垂直分布[J]. 微生物学报, 2020, 60(9): 1865−1881.Sun Hao, Tan Siyin, Liang Jinchang, et al. Horizontal and vertical distribution of dimethylsulfoniopropionate (DMSP) producing and catabolizing bacteria in the East China Sea[J]. Acta Microbiologica Sinica, 2020, 60(9): 1865−1881. [44] Zhang Yanying, Ling Juan, Yang Qingsong, et al. The functional gene composition and metabolic potential of coral-associated microbial communities[J]. Scientific Reports, 2015, 5: 16191. doi: 10.1038/srep16191 [45] Rosano P M, Leite D C A, Duarte G A S, et al. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation[J]. The ISME Journal, 2019, 13(4): 921−936. doi: 10.1038/s41396-018-0323-6 [46] Raina J B, Clode P L, Cheong S, et al. Subcellular tracking reveals the location of dimethylsulfoniopropionate in microalgae and visualises its uptake by marine bacteria[J]. eLife Sciences, 2017, 6: e23008. doi: 10.7554/eLife.23008 [47] Ritchie K B. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria[J]. Marine Ecology Progress Series, 2006, 322: 1−14. doi: 10.3354/meps322001 [48] Raina J B, Tapiolas D, Motti C A, et al. Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals[J]. PeerJ, 2016, 4 (6242): e2275. doi: 10.7717/peerj.2275 -




 下载:
下载: