Genetic diversity of Parasesarma affine from the South China Sea based on mitochondrial DNA COⅠ gene
-
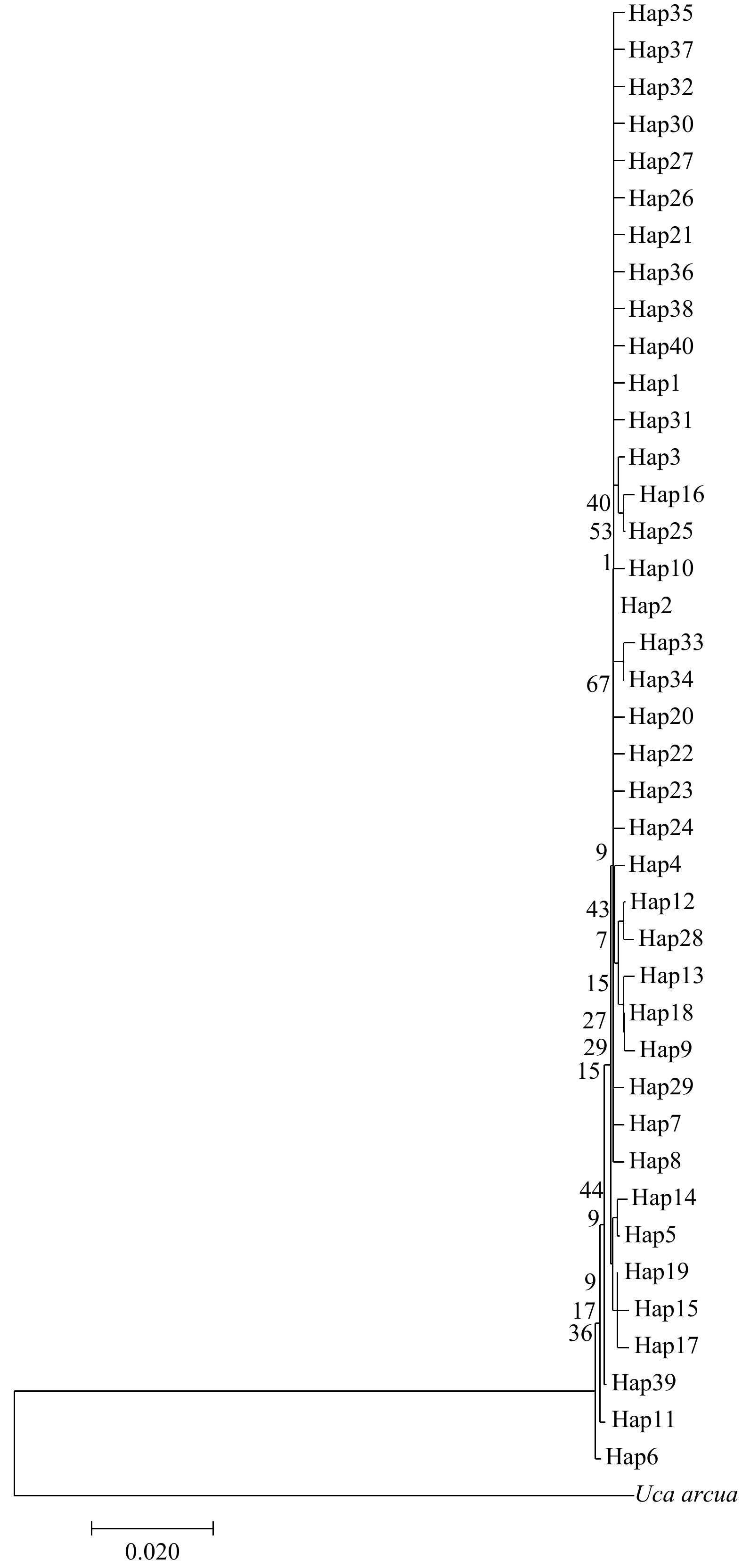
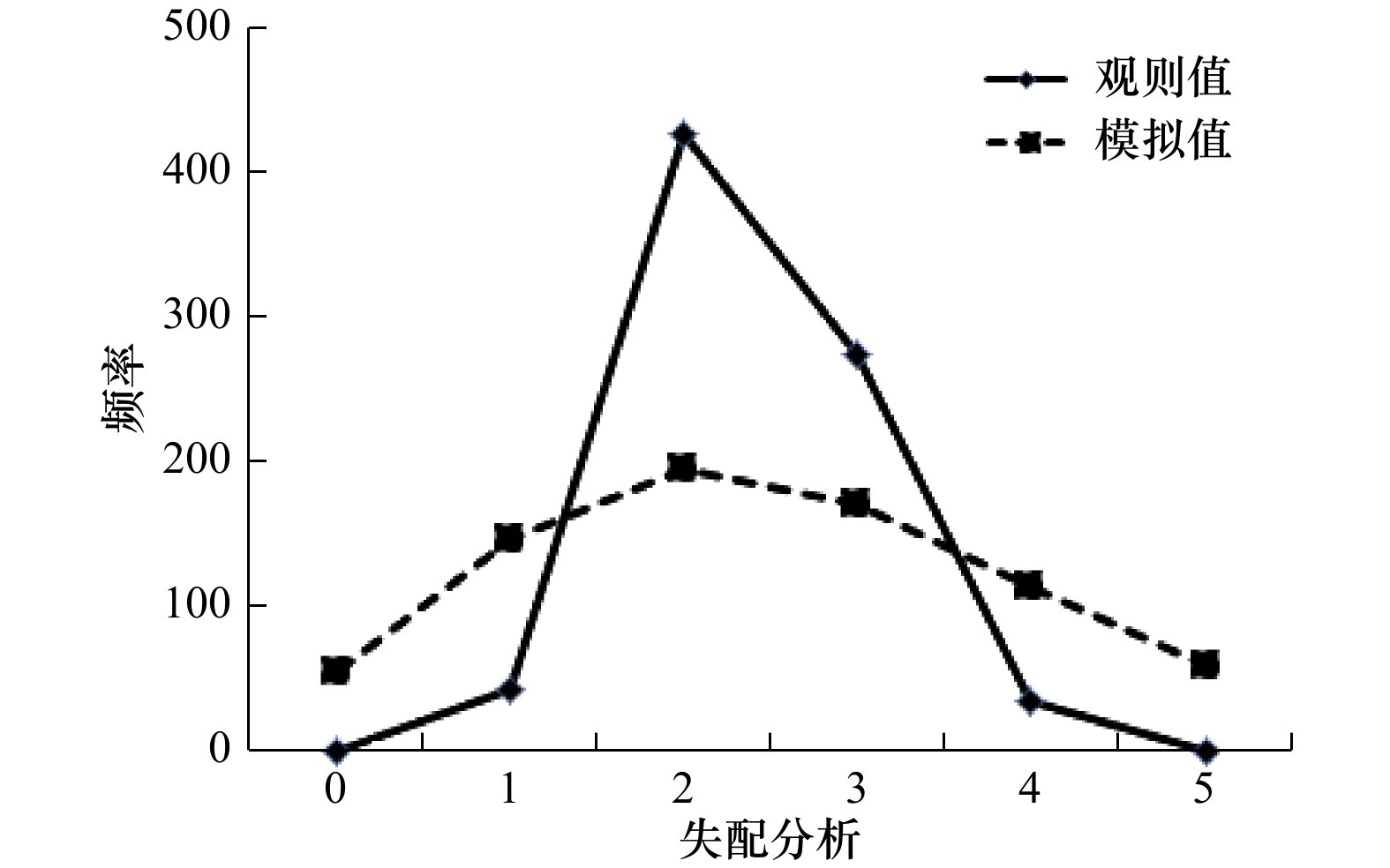
摘要: 为了研究我国南部沿海近亲拟相手蟹(Parasesarma affine)的群体遗传结构,本研究对12个地理群体共222个个体的线粒体DNA的细胞色素氧化酶亚基Ⅰ (COⅠ)基因片段进行了分析。结果表明,612 bp的COⅠ基因片段检测到34个变异位点,共定义了40个单倍型,其中Hap2为12个群体的共享单倍型,占个体总数的69.81%。总群体的单倍型多样性水平Hd为0.508 9,核苷酸多样性水平Pi为0.001 126,表现出中等水平的Hd和低水平的Pi。单倍型邻接发育树和单倍型中介网络图没有形成明显的地理系谱结构。近亲拟相手蟹群体内的遗传距离为0.000 36~0.001 73,群体间的遗传距离为0.000 48~0.001 72。群体间的遗传分化系数(Fst)和分子方差分析(AMOVA)结果表明,近亲拟相手蟹群体遗传分化水平低,其变异主要来自群体内。中性检验和核苷酸不配对分布结果提示,近亲拟相手蟹近期经历了群体扩张事件,扩张时间大约发生于5.1万年前的更新世晚期。研究表明,较长的幼虫浮游期以及海洋环境中缺少影响群体扩散的屏障可能是近亲拟相手蟹各地理群体间能进行广泛的基因交流,从而表现出较低的遗传分化水平的重要原因,更新世的剧烈气候变迁亦可能对其群体的遗传结构和分布格局产生影响。研究结果为近亲拟相手蟹自然资源的保护及合理开发利用提供了一定理论依据。Abstract: In this study, the population genetic diversity and structure of Parasesarma affine in the South China Sea were analyzed based on 222 individuals of twelve populations using Cytochrome Oxidase Ⅰ (COⅠ) sequence. A 612 bp fragment of COⅠ gene were sequenced, from which 34 polymorphic sites were tested and 40 haplotypes were defined. The most frequent haplotype was Hap2 with the highest frequency of 69.81%, which was shared in all twelve populations. Total haplotype diversity (Hd) and nucleotide diversity (Pi) were 0.508 9, 0.001 126, respectively, which represented a moderate level of haplotype diversity and a low level of nucleotide diversity. No clustering corresponding to sampling localities was found in neighbor-joining tree and haplotype network. The genetic distance ranged from 0.000 36 to 0.001 73 within populations and from 0.000 48 to 0.001 72 between populations. Genetic fixation index (Fst) and analysis of molecular variance (AMOVA) indicated that the genetic variance mainly came from individuals within populations, and a low level of genetic differentiation among twelve populations. Both neutral test and mismatch-distribution analysis implied that the populations of P. affine had a recent population expansion event that occurred in about 51 000 years ago in the late Pleistocene epoch. In summary, the longer time of the planktonic larval phase and the lack of evident geographical barriers in the marine realm might be major reasons for that P. affine could carry out extensive gene flow and possessed a low level of genetic differentiation among all twelve populations. In addition, the severe climate change of pleistocene epoch might also have important effects on the genetic structure and patterns of distribution of P. affine populations. The research results could provide a theoretical basis for the conservation and utilization of P. affine.
-
Key words:
- Parasesarma affine /
- COⅠ gene /
- genetic diversity /
- genetic structure
-
表 1 中国南部沿海近亲拟相手蟹的采样信息
Tab. 1 The sampling information of P. affine from the South China Sea
群体 采样地点 经纬度 样本数 采集时间 LH 福建龙海 24°28′06.4″N,117°54′19.5″E 22 2012年9月 YX 福建云霄 23°55′15.3″N,117°25′01.5″E 22 2012年9月 ZH 广东珠海 22°23′45.4″N,113°38′37.8″E 22 2013年6月 YJ 广东阳江 21°46′12.4″N,111°44′49.8″E 14 2013年6月 LF 广东雷州附城镇 20°53′18.2″N,110°10′30.1″E 22 2012年10月 LQ 广东雷州企水镇 20°48′13.2″N,109°44′9.7″E 22 2012年10月 XW 广东徐闻 20°14′47.7″N,110°7′19.3″E 14 2012年10月 SK 广西合浦山口镇 21°29′55.5″N,109°45′41.0″E 10 2012年7月 BH 广西北海 21°24′51.3″N,109°10′25.2″E 21 2012年7月 FCG 广西防城港 21°37′06.2″N,108°14′07.9″E 22 2012年7月 YP 海南洋浦 19°46′08.8″N,109°15′15.0″E 22 2012年10月 DF 海南东方 19°12′58.4″N,108°38′15.2″E 9 2012年10月 表 2 近亲拟相手蟹不同群体COⅠ基因片段的碱基组成和遗传多样性
Tab. 2 The base composition and genetic diversity parameters of COⅠ gene fragments among different population of P. affine
群体 碱基组成/% 单倍型多样性(Hd) 核苷酸多样性(Pi) T C A G A+T C+G LH 37.25 17.16 28.94 16.64 66.19 23.9 0.259 7±0.120 2 0.000 594±0.000 664 YX 37.24 17.18 28.92 16.66 66.16 33.84 0.593 1±0.116 7 0.001 146±0.001 003 ZH 37.25 17.16 28.91 16.67 66.16 33.93 0.545 5±0.127 6 0.001 040±0.000 941 YJ 37.22 17.18 28.94 16.65 66.16 33.83 0.494 5±0.150 6 0.000 898±0.000 880 LF 37.23 17.18 28.91 16.67 66.14 33.85 0.601 7±0.120 6 0.001 174±0.001 019 LQ 37.25 17.16 28.92 16.66 66.17 33.82 0.710 0±0.106 4 0.001 726±0.001 326 XW 37.24 17.17 28.94 16.64 66.18 33.81 0.395 6±0.158 8 0.000 700±0.000 753 SK 37.25 17.16 28.92 16.67 66.17 33.83 0.377 8±0.181 3 0.000 654±0.000 745 BH 37.25 17.17 28.94 16.64 66.19 33.81 0.681 0±0.113 1 0.001 696±0.001 313 FCG 37.28 17.14 18.96 16.62 56.24 33.76 0.541 1±0.125 3 0.001 712±0.001 319 YP 37.24 17.17 28.94 16.65 66.18 33.82 0.411 3±0.130 8 0.000 891±0.000 852 DF 37.27 17.14 28.92 16.67 66.19 33.81 0.222 2±0.166 2 0.000 362±0.000 534 总群体 37.25 17.17 28.93 16.65 66.18 33.82 0.508 9±0.041 9 0.001 126±0.000 951 表 3 近亲拟相手蟹12个群体单倍型分布情况
Tab. 3 Haplotype distribution of P. affine in twelve populations
单倍型 群体 合计 基因登记号 LH YX ZH YJ LF LQ XW SK BH FCG YP DF Hap1 − − − − − 1 − − 2 − − − 3 MT310974 Hap2 19 14 15 10 14 12 11 8 12 15 17 8 155 MT310975 Hap3 1 3 − 2 2 1 1 1 1 1 1 − 14 MT310976 Hap4 − − − − − − − − 1 − − − 1 MT310977 Hap5 − − − − − − − − 1 − − − 1 MT310978 Hap6 − − − − − − − − 1 − − − 1 MT310979 Hap7 − − − − − − − − 1 − 1 − 2 MT310980 Hap8 − − − − − − − − 1 − − − 1 MT310981 Hap9 − − − − − − − − 1 − − − 1 MT310982 Hap10 − − − − − − − − − − − 1 1 MT310983 Hap11 − − − − − − − − − 1 − − 1 MT310984 Hap12 − − − − − − 1 − − 1 − − 2 MT310985 Hap13 − − − − − − − − − 2 − − 2 MT310986 Hap14 − − − − − − − − − 1 − − 1 MT310987 Hap15 − − − − − − − − − 1 − − 1 MT310988 Hap16 − − − − − 1 − − − − − − 1 MT310989 Hap17 − − − − − 1 − − − − − − 1 MT310990 Hap18 − 1 − − − 1 − − − − − − 2 MT310991 Hap19 − − − − − 2 − − − − − − 2 MT310992 Hap20 − − 1 − − 1 − − − − − − 2 MT310993 Hap21 − − − − − 1 − − − − − − 1 MT310994 Hap22 − − − − 1 1 1 − − − 1 − 4 MT310995 Hap23 − − − − 1 − − − − − − − 1 MT310996 Hap24 − − − − 1 − − − − − − − 1 MT310997 Hap25 − − − − 1 − − − − − − − 1 MT310998 Hap26 − − − − 1 − − − − − − − 1 MT310999 Hap27 − − − − 1 − − − − − − − 1 MT311000 Hap28 1 − − − − − − − − − − − 1 MT311001 Hap29 1 − − 1 − − − − − − − − 2 MT311002 Hap30 − 1 1 − − − − 1 − − − − 3 MT311003 Hap31 − − − 1 − − − − − − − − 1 MT311004 Hap32 − − − − − − − − − − 1 − 1 MT311005 Hap33 − − − − − − − − − − 1 − 1 MT311006 Hap34 − 1 1 − − − − − − − − − 2 MT311007 Hap35 − 1 − − − − − − − − − − 1 MT311008 Hap36 − 1 − − − − − − − − − − 1 MT311009 Hap37 − − 1 − − − − − − − − − 1 MT311010 Hap38 − − 1 − − − − − − − − − 1 MT311011 Hap39 − − 1 − − − − − − − − − 1 MT311012 Hap40 − − 1 − − − − − − − − − 1 MT311013 合计 22 22 22 14 22 22 14 10 21 22 22 9 222 − 注:−代表未检测到单倍型。 表 4 基于COⅠ基因序列的近亲拟相手蟹群体间遗传分化系数Fst值(对角线下)和群体内(对角线) 及群体间(对角线上)遗传距离
Tab. 4 F-statistics (below diagonal) within populations and genetic distance within (diagonal) and between (above diagonal) populations of P. affine based on COⅠ gene
LH YX ZH YJ LF LQ XW SK BH FCG YP DF LH 0.000 60 0.000 87 0.000 82 0.000 73 0.000 88 0.001 17 0.000 63 0.000 61 0.001 14 0.001 16 0.000 74 0.000 48 YX −0.006 70 0.001 15 0.001 10 0.001 00 0.001 15 0.001 44 0.000 91 0.000 86 0.001 43 0.001 46 0.001 02 0.000 78 ZH 0.000 00 0.007 01 0.001 04 0.000 99 0.001 12 0.001 41 0.000 87 0.000 83 0.001 37 0.001 41 0.000 96 0.000 70 YJ −0.014 01 −0.027 17 0.016 03 0.000 90 0.001 02 0.001 32 0.000 79 0.000 75 0.001 30 0.001 34 0.000 89 0.000 65 LF −0.007 33 −0.010 64 0.006 35 −0.020 15 0.001 18 0.001 45 0.000 91 0.000 89 0.001 44 0.001 48 0.001 02 0.000 78 LQ 0.006 88 0.001 56 0.015 34 −0.005 18 −0.003 56 0.001 73 0.001 21 0.001 19 0.001 72 0.001 70 0.001 32 0.001 08 XW −0.032 20 −0.017 28 −0.005 01 −0.019 64 −0.032 32 −0.013 10 0.000 70 0.000 65 0.001 20 0.001 22 0.000 78 0.000 53 SK −0.022 01 −0.055 04 −0.028 66 −0.042 29 −0.038 04 −0.023 05 −0.036 58 0.000 65 0.001 17 0.001 21 0.000 76 0.000 51 BH −0.004 57 0.000 93 0.001 05 −0.004 57 0.000 90 0.000 39 −0.012 07 −0.026 96 0.001 70 0.001 71 0.001 29 0.001 04 FCG 0.007 20 0.015 73 0.020 38 0.014 93 0.019 66 −0.014 23 0.000 47 −0.006 72 0.000 40 0.001 72 0.001 33 0.001 08 YP −0.009 17 −0.005 71 −0.007 04 −0.003 57 −0.012 93 0.006 11 −0.029 92 −0.026 99 −0.004 28 0.021 51 0.000 89 0.000 63 DF −0.015 02 −0.005 80 −0.027 68 0.009 76 −0.022 01 −0.013 50 −0.012 41 −0.003 32 −0.028 18 −0.007 85 −0.024 65 0.000 36 表 5 近亲拟相手蟹12个群体的AMOVA分析
Tab. 5 Analysis of molecular variance (AMOVA) for the twelve P. affine populations
变异来源 自由度 平方和 变异组成 变异百分比/% 固定值FST 组群间 11 3.526 −0.001 38 Va −0.4 −0.004 02 (p=0.757 6) 种群内 210 72.654 0.345 97 $^{{\rm{V_b}}} $ 100.4 合计 221 76.18 0.344 59 100 注:Va 表示组群间方差;Vb 表示组群内群体间方差。 表 6 近亲拟相手蟹12个群体的中性检验和核苷酸不配对分布
Tab. 6 Neutral testes and mismatch distributions of nucleotide in twelve P. affine populations
群体 Tajima’s D Fu’s Fs 核苷酸不配对分布 D p Fs p τ SSD Rg LH −1.877 63 0.011 00 −2.205 61 0.006 00 3.000 00 0.002 13 0.343 72 YX −1.778 56 0.019 00 −4.749 13 0.000 00 0.875 00 0.017 16 0.159 63 ZH −2.139 60 0.001 00 −6.978 93 0.000 00 0.783 20 0.011 77 0.140 50 YJ −1.278 26 0.085 00 −1.727 37 0.012 00 0.683 59 0.012 90 0.155 30 LF −2.002 53 0.007 00 −6.347 57 0.000 00 0.894 53 0.016 49 0.156 56 LQ −1.917 76 0.008 00 −7.818 78 0.000 00 1.166 02 0.005 87 0.092 15 XW −1.670 53 0.026 00 −2.288 11 0.002 00 0.505 86 0.005 27 0.168 22 SK −1.400 85 0.077 00 −1.163 94 0.036 00 0.490 23 0.005 79 0.182 72 DG −2.162 55 0.004 00 −6.321 53 0.000 00 1.115 23 0.001 64 0.067 10 FCG −1.715 70 0.018 00 −3.162 46 0.007 00 2.312 50 0.055 27 0.243 44 YP −2.071 98 0.002 00 −4.121 61 0.000 00 0.705 08 0.000 06 0.131 67 DF −1.088 23 0.191 00 −0.263 48 0.154 00 2.929 69 0.306 72 0.358 02 合计 −2.497 80 0.000 00 −29.929 97 0.000 00 0.714 84 0.000 00 0.950 00 -
[1] 刘瑞玉. 中国海洋生物名录[M]. 北京: 科学出版社, 2008: 801−802.Liu Ruiyu. Checklist of Marine Biota of China Seas[M]. Beijing: Science Press, 2008: 801−802. [2] 杨明柳, 徐敬明, 吴斌, 等. 北部湾红树林蟹类多样性初步研究[J]. 四川动物, 2014, 33(3): 347−352, 357.Yang Mingliu, Xu Jingming, Wu Bin, et al. A preliminary study on crabs diversity for Beibu Gulf mangrove[J]. Sichuan Journal of Zoology, 2014, 33(3): 347−352, 357. [3] Chen Guangcheng, Ye Yong. Leaf consumption by Sesarma plicata in a mangrove forest at Jiulongjiang Estuary, China[J]. Marine Biology, 2008, 154(6): 997−1007. doi: 10.1007/s00227-008-0990-3 [4] Lee S Y. Ecological role of grapsid crabs in mangrove ecosystems: a review[J]. Marine and Freshwater Research, 1998, 49(4): 335−343. doi: 10.1071/MF97179 [5] Lee S Y, Kwok P W. The importance of mangrove species association to the population biology of the sesarmine crabs Parasesarma affinis and Perisesarma bidens[J]. Wetlands Ecology and Management, 2002, 10(3): 215−226. doi: 10.1023/A:1020175729972 [6] Chen G C, Lu Changyi, Li Rong, et al. Effects of foraging leaf litter of Aegiceras corniculatum (Ericales, Myrsinaceae) by Parasesarma plicatum (Brachyura, Sesarmidae) crabs on properties of mangrove sediment: a laboratory experiment[J]. Hydrobiologia, 2016, 763(1): 125−133. doi: 10.1007/s10750-015-2367-1 [7] Wang Zhengfei, Wang Ziqian, Shi Xuejia, et al. Complete mitochondrial genome of Parasesarma affine (Brachyura: Sesarmidae): gene rearrangements in Sesarmidae and phylogenetic analysis of the Brachyura[J]. International Journal of Biological Macromolecules, 2018, 118: 31−40. doi: 10.1016/j.ijbiomac.2018.06.056 [8] 杨明柳, 徐敬明, 吴斌, 等. 基于形态和16S rRNA基因的拟相手蟹分类研究[J]. 海洋学报, 2016, 38(8): 62−72.Yang Mingliu, Xu Jingming, Wu Bin, et al. Taxonomic study on the genus Parasesarma based on the morphology characteristics and mtDNA 16S rRNA gene[J]. Haiyang Xuebao, 2016, 38(8): 62−72. [9] 陈光程, 余丹, 叶勇, 等. 红树林植被对大型底栖动物群落的影响[J]. 生态学报, 2013, 33(2): 327−336. doi: 10.5846/stxb201111091699Chen Guangcheng, Yu Dan, Ye Yong, et al. Impacts of mangrove vegetation on macro-benthic faunal communities[J]. Acta Ecologica Sinica, 2013, 33(2): 327−336. doi: 10.5846/stxb201111091699 [10] 卢元平, 徐卫华, 张志明, 等. 中国红树林生态系统保护空缺分析[J]. 生态学报, 2019, 39(2): 684−691.Lu Yuanping, Xu Weihua, Zhang Zhiming, et al. Gap analysis of mangrove ecosystem conservation in China[J]. Acta Ecologica Sinica, 2019, 39(2): 684−691. [11] Moritz C, Dowling T E, Brown W M. Evolution of animal mitochondrial DNA: relevance for population biology and systematics[J]. Annual Review of Ecology and Systematics, 1987, 18(1): 269−292. doi: 10.1146/annurev.es.18.110187.001413 [12] Xu Jingming, Yang Fan, Sun Shichun. Population genetic structure and phylogeography of the mud-flat crabs Helice tientsinensis and Helice latimera along the coast of China seas based on mitochondrial DNA[J]. African Journal of Biotechnology, 2012, 11(16): 3738−3750. [13] Ma Hongyu, Ma Chunyan, Ma Lingbo. Population genetic diversity of mud crab (Scylla paramamosain) in Hainan Island of China based on mitochondrial DNA[J]. Biochemical Systematics and Ecology, 2011, 39(4/6): 434−440. [14] Zhou Haolang, Xu Jingming, Yang Mingliu, et al. Population genetic diversity of sesarmid crab (Perisesarma bidens) in China based on mitochondrial DNA[J]. Mitochondrial DNA Part A, 2016, 27(5): 3255−3262. doi: 10.3109/19401736.2015.1015002 [15] 王景, 张凤英, 蒋科技, 等. 基于线粒体COⅠ基因序列的三疣梭子蟹东海区群体遗传多样性分析[J]. 海洋渔业, 2015, 37(2): 114−121. doi: 10.3969/j.issn.1004-2490.2015.02.003Wang Jing, Zhang Fengying, Jiang Keji, et al. Genetic diversity of Portunus trituberculatus based on the Mitochondrial Cytochrome Oxidase Subunit I Sequence from the East China Sea[J]. Marine Fisheries, 2015, 37(2): 114−121. doi: 10.3969/j.issn.1004-2490.2015.02.003 [16] Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates[J]. Molecular Marine Biology and Biotechnology, 1994, 3(5): 294−299. [17] Burland T G. DNASTAR’s Lasergene sequence analysis software[M]//Misener S, Krawetz S A. Bioinformatics Methods and Protocols. Totowa, NJ: Humana Press, 2000: 71−91. [18] Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data[J]. Bioinformatics, 2009, 25(11): 1451−1452. doi: 10.1093/bioinformatics/btp187 [19] Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis[J]. Evolutionary Bioinformatics Online, 2005, 1: 47−50. [20] Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Molecular Biology and Evolution, 2016, 33(7): 1870−1874. doi: 10.1093/molbev/msw054 [21] Polzin T, Daneshmand S V. On Steiner trees and minimum spanning trees in hypergraphs[J]. Operations Research Letters, 2003, 31(1): 12−20. doi: 10.1016/S0167-6377(02)00185-2 [22] Fu Y X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection[J]. Genetics, 1997, 147(2): 915−925. [23] Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism[J]. Genetics, 1989, 123(3): 585−595. [24] Schubart C D, Diesel R, Hedges S B. Rapid evolution to terrestrial life in Jamaican crabs[J]. Nature, 1998, 393(6683): 363−365. doi: 10.1038/30724 [25] Daniels S R, Stewart B A, Gouws G, et al. Phylogenetic relationships of the southern African freshwater crab fauna (Decapoda: Potamonautidae: Potamonautes) derived from multiple data sets reveal biogeographic patterning[J]. Molecular Phylogenetics and Evolution, 2002, 25(3): 511−523. doi: 10.1016/S1055-7903(02)00281-6 [26] Ragionieri L, Cannicci S, Schubart C D, et al. Gene flow and demographic history of the mangrove crab Neosarmatium meinerti: a case study from the western Indian Ocean[J]. Estuarine, Coastal and Shelf Science, 2010, 86(2): 179−188. doi: 10.1016/j.ecss.2009.11.002 [27] 吴晓雯, 余海, 陈青萍, 等. 基于COⅠ基因对温州沿海岛屿疣荔枝螺遗传多样性研究[J]. 动物学杂志, 2019, 54(2): 254−269.Wu Xiaowen, Yu Hai, Chen Qingping, et al. Genetic diversity of Reishia clavigera on the coastal islands of wenzhou based on COⅠ gene[J]. Chinese Journal of Zoology, 2019, 54(2): 254−269. [28] 金丹璐, 娄剑锋, 高心明, 等. 基于线粒体COⅠ基因序列的东南沿海可口革囊星虫遗传多样性分析[J]. 水生生物学报, 2017, 41(6): 1257−1264. doi: 10.7541/2017.156Jin Danlu, Lou Jianfeng, Gao Xinming, et al. The genetic diversity of Phascolosoma esculenta in the coastal zone of south-eastern China based on sequence analysis of mitochondrial CO I gene[J]. Acta Hydrobiologica Sinica, 2017, 41(6): 1257−1264. doi: 10.7541/2017.156 [29] 宋忠魁, 李梦芸, 聂振平, 等. 北部湾拟穴青蟹(Scylla paramamosain)群体遗传结构及其扩张分析[J]. 海洋与湖沼, 2012, 43(4): 828−836. doi: 10.11693/hyhz201204021021Song Zhongkui, Li Mengyun, Nie Zhenping, et al. Population genetic structure and expansion analysis of green mud crab (Scylla paramamosain) from Beibu Gulf[J]. Oceanologia et Limnologia Sinica, 2012, 43(4): 828−836. doi: 10.11693/hyhz201204021021 [30] Grant W S, Bowen B W. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation[J]. Journal of Heredity, 1998, 89(5): 415−426. doi: 10.1093/jhered/89.5.415 [31] Silva I C, Mesquita N, Paula J. Genetic and morphological differentiation of the mangrove crab Perisesarma guttatum (Brachyura: Sesarmidae) along an East African latitudinal gradient[J]. Biological Journal of the Linnean Society, 2010, 99(1): 28−46. [32] Li Jiji, Ye Yingying, Wu Changwen, et al. Genetic variation of Mytilus coruscus Gould (Bivalvia: Mytilidae) populations in the East China Sea inferred from mtDNA COI gene sequence[J]. Biochemical Systematics and Ecology, 2013, 50: 30−38. doi: 10.1016/j.bse.2013.03.033 [33] Cassone B J, Boulding E G. Genetic structure and phylogeography of the lined shore crab, Pachygrapsus crassipes, along the northeastern and western Pacific coasts[J]. Marine Biology, 2006, 149(2): 213−226. doi: 10.1007/s00227-005-0197-9 [34] Slatkin M, Hudson R R. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations[J]. Genetics, 1991, 129(2): 555−562. [35] Hewitt G. The genetic legacy of the Quaternary ice ages[J]. Nature, 2000, 405(6789): 907−913. doi: 10.1038/35016000 [36] Palumbi S R. Genetic divergence, reproductive isolation, and marine speciation[J]. Annual Review of Ecology and Systematics, 1994, 25(1): 547−572. doi: 10.1146/annurev.es.25.110194.002555 [37] Skov M W, Hartnoll R G, Ruwa R K, et al. Marching to a different drummer: crabs synchronize reproduction to a 14-month lunar-tidal cycle[J]. Ecology, 2005, 86(5): 1164−1171. doi: 10.1890/04-0917 [38] Palumbi S R. Population genetics, demographic connectivity, and the design of marine reserves[J]. Ecological Applications, 2003, 13(S1): 146−158. [39] Krishnan T, Kannupandi T. Larval development of the mangrove crab Sesarma bidens (De Haan, 1853) in the laboratory (Brachyura: Grapsidae: Sesarminae)[J]. Mahasagar, 1987, 20(3): 171−181. [40] Silva I C, Mesquita N, Paula J. Lack of population structure in the fiddler crab Uca annulipes along an East African latitudinal gradient: genetic and morphometric evidence[J]. Marine Biology, 2010, 157(5): 1113−1126. doi: 10.1007/s00227-010-1393-9 [41] 沈浪, 陈小勇, 李媛媛. 生物冰期避难所与冰期后的重新扩散[J]. 生态学报, 2002, 22(11): 1983−1990. doi: 10.3321/j.issn:1000-0933.2002.11.026Shen Lang, Chen Xiaoyong, Li Yuanyuan. Glacial refugia and postglacial recolonization patterns of organisms[J]. Acta Ecologica Sinica, 2002, 22(11): 1983−1990. doi: 10.3321/j.issn:1000-0933.2002.11.026 [42] Hewitt G M. Genetic consequences of climatic oscillations in the Quaternary[J]. Philosophical Transactions of the Royal Society B: Biological Sciences, 2004, 359(1442): 183−195. doi: 10.1098/rstb.2003.1388 [43] Lambeck K, Esat T M, Potter E K. Links between climate and sea levels for the past three million years[J]. Nature, 2002, 419(6903): 199−206. doi: 10.1038/nature01089 [44] Laurenzano C, Costa T M, Schubart C D. Contrasting patterns of clinal genetic diversity and potential colonization pathways in two species of western Atlantic fiddler crabs[J]. PLoS One, 2016, 11(11): e0166518. doi: 10.1371/journal.pone.0166518 [45] Laurenzano C, Mantelatto F L M, Schubart C D. South American homogeneity versus Caribbean heterogeneity: population genetic structure of the western Atlantic fiddler crab Uca rapax (Brachyura, Ocypodidae)[J]. Journal of Experimental Marine Biology and Ecology, 2013, 449: 22−27. doi: 10.1016/j.jembe.2013.08.007 [46] 柯富士, 尤士骏, 黄素梅, 等. 中国小菜蛾群体遗传变异的时空动态[J]. 昆虫学报, 2019, 62(5): 624−633.Ke Fushi, You Shijun, Huang Sumei, et al. Spatiotemporal dynamics of genetic variation in populations of the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae), in China[J]. Acta Entomologica Sinica, 2019, 62(5): 624−633. [47] Okeyo W A, Saarman N P, Mengual M, et al. Temporal genetic differentiation in Glossina pallidipes tsetse fly populations in Kenya[J]. Parasites & Vectors, 2017, 10(1): 471. -





 下载:
下载: