Transcriptome analysis provides insights into the response of coelomocytes in polian vesicle and coelomic cavity of sea cucumber Apostichopus japonicus to evisceration
-
摘要: 刺参的体腔细胞大量存在于体腔液与水管系统内,广泛参与机体的营养输送、代谢以及免疫等多种功能。刺参吐脏时,体腔中体腔细胞近乎排尽,而后迅速恢复。波里氏囊是刺参吐脏后仅存的内脏器官,囊腔内体腔细胞在吐脏后也快速增加,表现出积极的响应。为研究探讨刺参吐脏后再生早期体腔细胞快速增加的作用与意义,本文分别对刺参吐脏后6 h与吐脏前波里氏囊腔和体腔中体腔细胞进行转录组测序,比较了波里氏囊腔与体腔中体腔细胞在吐脏胁迫下的基因表达变化。结果显示,波里氏囊腔内体腔细胞在吐脏前后有267个基因显著差异表达,差异基因大量富集至GO功能注释的酶催化活性亚类以及甘氨酸、丝氨酸与苏氨酸代谢等KEGG通路。体腔中体腔细胞在吐脏前后有922个基因显著差异表达,差异基因显著富集到GO功能注释的细胞黏附、生物黏附等亚类以及细胞外基质受体互作、转化生长因子-β与FoxO等KEGG通路。研究结果对于进一步研究刺参体腔细胞的功能及揭示刺参吐脏后的再生机制提供了重要基础。Abstract: Coelomocytes in Apostichopus japonicus, present in coelomic fluid and water-vascular system, are considered to participate in a variety of biological functions including nutrition transport, metabolism and immunity. In the process of evisceration, coelomocytes in coelom are nearly exhausted and then recovered in a short period. The polian vesicle, as the only remained internal organ after evisceration, shows positive response that coelomocytes within it increased rapidly. Coelomocytes in coelom are nearly exhausted after evisceration, and then recovered quickly. To further investigate the function and significance of the rapid increase of coelomocytes in the early stage of regeneration after evisceration, transcriptome sequencing was performed for coelomocytes in polian vesicle and coelom of A. japonicus at 6 h after evisceration and pre-evisceration, respectively. The gene expression differences of coelomocytes in polian vesicle and coelom after evisceration were analyzed compared to those of healthy A. japonicus. These results showed that 267 genes were differentially expressed in coelomocytes of polian vesicle at 6 h after evisceration, and most of these genes were enriched into the enzyme catalytic activity subclasses according to GO functional annotation and glycine, serine and threonine metabolism pathway according to KEGG pathways annotation, respectively. Additionally, 922 differential genes were significantly expressed in coelomocytes of coelom at 6 h after evisceration, and these genes were enriched into cell adhesion and biological adhesion subclasses according to GO functional annotation and ECM-receptor interaction, TGF-β signaling pathway, FoxO signaling pathway according to KEGG pathways annotation, respectively. The results provide an important basis for further functional research and the regeneration mechanism of coelomocytes after evisceration in A. japonicus.
-
Key words:
- Apostichopus japonicus /
- coelomocytes /
- polian vesicle /
- evisceration /
- transcriptome
-
图 1 PC6 h组与PC0 h组(a)及CC6 h组与CC0 h组(b)的显著差异基因分布火山图
红点代表log2 (Fold change)>1和padj<0.05的上调基因;绿点表示log2 (Fold change)<−1和padj<0.05的下调基因;蓝点代表没有显著差异的基因
Fig. 1 Volcano plot significantly differential expression genes distribution between PC6 h and PC0 h (a) and between CC6 h and CC0 h (b)
Red points represent up-regulated genes with log2(Fold change)>1 and padj<0.05; green points represent down-regulated genes with log2(Fold change)<−1 and padj<0.05; blue points represent genes with no significant difference
图 2 GO富集分析中显著差异基因的功能分类
PC6 h和PC0 h间显著差异基因的GO分析 (a);CC6 h与CC0 h间显著差异基因的GO分析 (b)。x轴表示GO分析中的亚类;y轴表示差异基因的数量;蓝色和绿色分别代表上调基因和下调基因
Fig. 2 Functional categorization of significantly differential expression genes in Gene Ontology
GO analysis between PC6 h and PC0 h (a); GO analysis between between CC6 h and CC0 h (b). The x-axis shows the 2nd level term of Gene Ontology; the y-axis shows the number of DEGs; the blue and brown represent up- and down-regulated genes, respectively
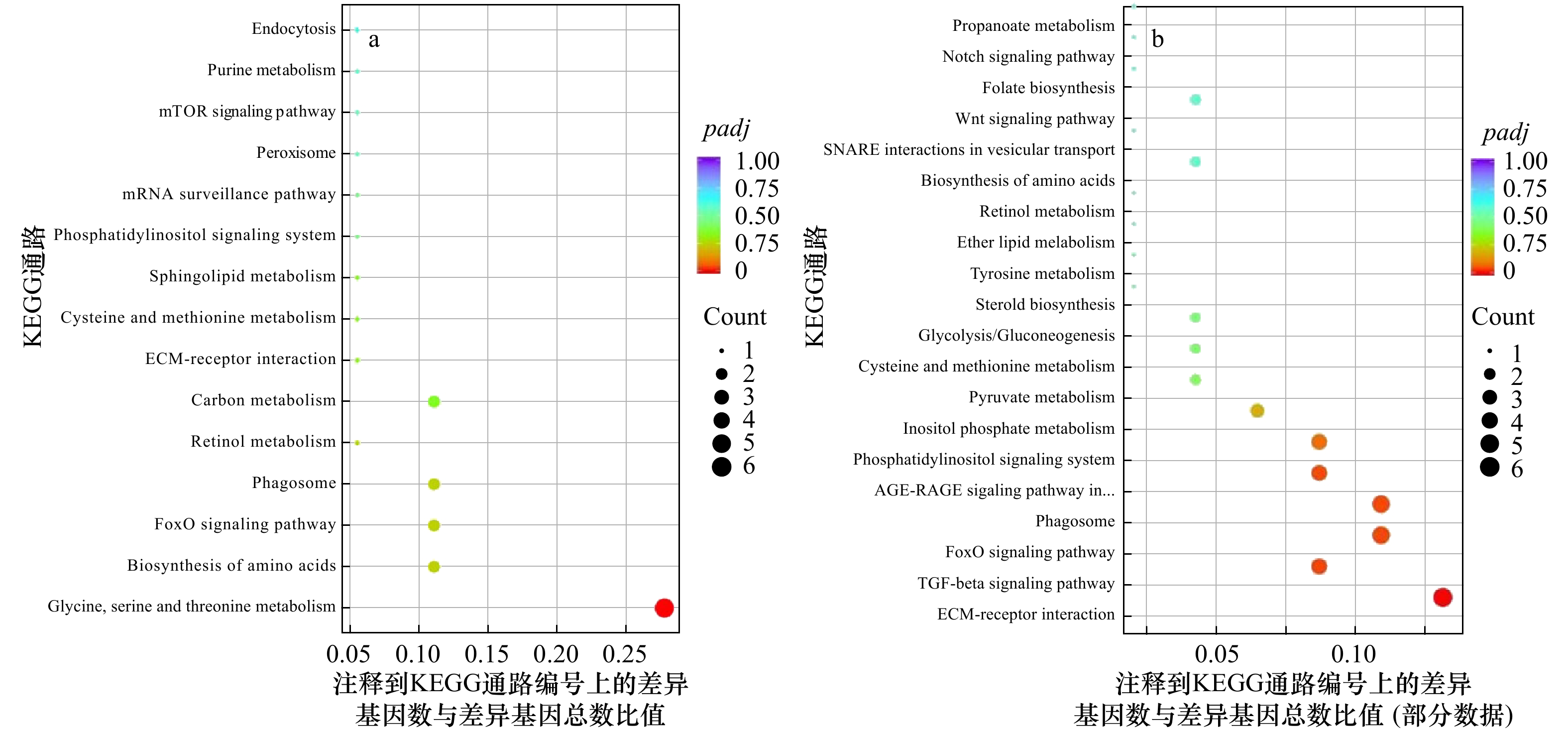
图 3 显著差异基因的KEGG通路富集分析
PC6 h与PC0 h间显著差异基因的KEGG通路富集 (a);CC6 h和CC0 h间显著差异基因的KEGG通路富集 (b)。圆点的颜色表示padj值,圆点的大小表示注释到KEGG通路上的基因数
Fig. 3 KEGG pathway enrichment of significantly differential expression genes
KEGG pathway enrichment between PC6 h and PC0 h (a); KEGG pathway enrichment between CC6 h and CC0 h (b). The color of the dot represents padj and size of the dot represents the number of DEGs mapped to the reference pathways
表 1 用qRT-PCR验证所用引物序列
Tab. 1 The sequence of primers used for qRT-PCR validation
基因名称 上游引物序列 下游引物序列 ADAMTS9 TGTGATGCTGGTGATTGCTTA GCCTTCGTCTTCGGCTTTA MMP16 TGGAAGTGGAATCGGTGGT GGATGTAAAGCGAAGTTAGGGT RARB GCTCCAACCAGCACCTACCTT AAACCCTTACACCCTTCGCA SLC6A7 AAGTCATCGGGCAAGGTCG GAAACAAGCAAAGCATCTCGGT FCGBP GAAGAACATCCTTGCCCCG GACATCATACACGCAGTCATCATAG PSAT1 GTAATGAGGGATTGGAAAAGCA ACGCAACCCACCGACAGA 表 2 12个文库的基本信息
Tab. 2 The basic characteristic of reads in the 12 libraries
样本 原始测序数据 过滤后数据 过滤后碱基数/Gb Q20值/% Q30值/% 比对率/% PC0 h-1 58 695 018 57 363 038 8.60 97.10 92.31 68.25 PC0 h-2 56 482 646 55 367 354 8.31 97.04 92.24 66.83 PC0 h-3 49 941 732 48 766 610 7.31 96.97 92.03 66.06 PC6 h-1 52 553 492 51 358 414 7.70 97.05 92.24 68.17 PC6 h-2 49 356 694 48 272 958 7.24 97.08 92.27 67.93 PC6 h-3 48 395 496 47 518 448 7.13 97.13 92.38 68.12 CC0 h-1 60 069 216 58 262 406 8.74 95.56 89.17 62.47 CC0 h-2 43 214 102 42 043 190 6.31 95.00 87.97 63.08 CC0 h-3 41 985 400 40 917 640 6.09 94.87 87.93 61.10 CC6 h-1 45 151 132 43 809 166 6.57 95.36 88.82 64.00 CC6 h-2 43 699 300 42 464 320 6.37 95.36 88.80 64.88 CC6 h-3 45 650 930 44 497 710 6.67 95.89 89.84 61.13 注:PC0 h代表吐脏前波里氏囊腔内体腔细胞;PC6 h代表吐脏后6 h波里氏囊腔内体腔细胞;CC0 h代表吐脏前体腔中体腔细胞;CC6 h代表吐脏后6 h体腔中体腔细胞。 表 3 显著富集通路中的差异表达基因
Tab. 3 The differential expression genes in significant enrichment pathways
基因名称 基因描述 表达倍数 Glycine,serine and threonine metabolism PSAT1 phosphoserine aminotransferase isoform X1 1.71 GNMT glycine N-methyltransferase 1.79 SARDH Sarcosine dehydrogenase, mitochondrial 1.72 BHMT betaine-homocysteine S-methyltransferase 1 1.48 ECM-receptor interaction COL4A5 collagen alpha-5(IV) chain-like isoform X2 2.71 THBS-1 thrombospondin-1 2.49 COL9A2 collagen alpha-2(IX) chain 3.37 HSPG2 basement membrane-specific heparan sulfate proteoglycan core protein 2.58 COMP cartilage oligomeric matrix protein 1.51 FoxO signaling pathway PCK1 Phosphoenolpyruvate carboxykinase 2.40 GRB2 growth factor receptor-bound protein 2 1.54 NLK2 serine/threonine-protein kinase NLK2 isoform X1 1.91 Cyclin B cyclin B −2.06 PLK polo-like kinase −2.20 TGF-beta signaling pathway FST follistatin isoform X2 3.00 -
[1] Dornbos S Q. Evolutionary palaeoecology of early epifaunal echinoderms: response to increasing bioturbation levels during the Cambrian radiation[J]. Palaeogeography, Palaeoclimatology, Palaeoecology, 2006, 237(2/4): 225−239. [2] Yang Hongsheng, Hamel J F, Mercier A. The Sea Cucumber Apostichopus japonicus: History, Biology and Aquaculture[M]. Boston, USA: Academic Press, 2015. [3] Li Qiang, Ren Yuan, Luan Linlin, et al. Localization and characterization of hematopoietic tissues in adult sea cucumber, Apostichopus japonicus[J]. Fish & Shellfish Immunology, 2019, 84: 1−7. [4] Li Qiang, Qi Ruirong, Wang Yi’nan, et al. Comparison of cells free in coelomic and water-vascular system of sea cucumber, Apostichopus japonicus[J]. Fish & Shellfish Immunology, 2013, 35(5): 1654−1657. [5] Ren Yuan, Zhang Jialin, Wang Yi’nan, et al. Non-specific immune factors differences in coelomic fluid from Polian vesicle and coelom of Apostichopus japonicus, and their early response after evisceration[J]. Fish & Shellfish Immunology, 2020, 98: 160−166. [6] Shukalyuk A I, Dolmatov L Y. Regeneration of the digestive tube in the holothurian Apostichopus japonicus after evisceration[J]. Russian Journal of Marine Biology, 2001, 27(3): 168−173. doi: 10.1023/A:1016717502616 [7] 王霞, 李霞. 仿刺参消化道的再生形态学与组织学[J]. 大连水产学院学报, 2007, 22(5): 340−346.Wang Xia, Li Xia. The morphological and histological observation of regeneration of alimentary tract in sea cucumber Apostichopus japonicus[J]. Journal of Dalian Fisheries University, 2007, 22(5): 340−346. [8] Sun Li’na, Xu Dongxue, Xu Qinzeng, et al. iTRAQ reveals proteomic changes during intestine regeneration in the sea cucumber Apostichopus japonicus[J]. Comparative Biochemistry and Physiology Part D: Genomics and Proteomics, 2017, 22: 39−49. doi: 10.1016/j.cbd.2017.02.004 [9] Li Qiang, Ren Yuan, Liang Chunlei, et al. Regeneration of coelomocytes after evisceration in the sea cucumber, Apostichopus japonicus[J]. Fish & Shellfish Immunology, 2018, 76: 266−271. [10] Liao Kuangming, Chao T B, Tian Yufeng, et al. Overexpression of the PSAT1 gene in nasopharyngeal carcinoma is an indicator of poor prognosis[J]. Journal of Cancer, 2016, 7(9): 1088−1094. doi: 10.7150/jca.15258 [11] Yang Yi, Wu Jueheng, Cai Junchao, et al. PSAT1 regulates cyclin D1 degradation and sustains proliferation of non-small cell lung cancer cells[J]. International Journal of Cancer, 2015, 136(4): E39−E50. doi: 10.1002/ijc.29150 [12] Frantz C, Stewart K M, Weaver V M. The extracellular matrix at a glance[J]. Journal of Cell Science, 2010, 123(24): 4195−4200. doi: 10.1242/jcs.023820 [13] Badylak S F. The extracellular matrix as a scaffold for tissue reconstruction[J]. Seminars in Cell & Developmental Biology, 2002, 13(5): 377−383. [14] Quinones J L, Rosa R, Ruiz D L, et al. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber Holothuria glaberrima[J]. Developmental Biology, 2002, 250(1): 181−197. doi: 10.1006/dbio.2002.0778 [15] García-Arrarás J E, Estrada-Rodgers L, Santiago R, et al. Cellular mechanisms of intestine regeneration in the sea cucumber, Holothuria glaberrima Selenka (Holothuroidea: Echinodermata)[J]. Journal of Experimental Zoology, 1998, 281(4): 288−304. doi: 10.1002/(SICI)1097-010X(19980701)281:4<288::AID-JEZ5>3.0.CO;2-K [16] 孙丽娜. 仿刺参Apostichopus japonicas (Selenka)消化道再生的组织细胞特征与关键基因分析[D]. 青岛: 中国科学院海洋研究所, 2013.Sun Li’na. Histocytological events and analysis of key genes during intestine regeneration in sea cucumber Apostichopus japonicus (Selenka)[D]. Qingdao: Institute of Oceanology, Chinese Academy of Sciences, 2013. [17] Hetzel H R. Studies on holothurian coelomocytes. Ⅱ. The origin of coelomocytes and the formation of brown bodies[J]. The Biological Bulletin, 1965, 128(1): 102−111. doi: 10.2307/1539393 [18] Endean R. The coelomocytes of Holothuria leucospilota[J]. Journal of Cell Science, 1958, 99(45): 47−60. [19] Borgne A, Ostvold A C, Flament S, et al. Intra-M phase-promoting factor phosphorylation of Cyclin B at the prophase/metaphase transition[J]. Journal of Biological Chemistry, 1999, 274(17): 11977−11986. doi: 10.1074/jbc.274.17.11977 [20] Richardson H, Lew D J, Henze M, et al. Cyclin-B homologs in saccharomyces cerevisiae function in S phase and in G2[J]. Genes & Development, 1992, 6(11): 2021−2034. [21] Barr F A, Silljé H H W, Nigg E A. Polo-like kinases and the orchestration of cell division[J]. Nature Reviews Molecular Cell Biology, 2004, 5(6): 429−441. doi: 10.1038/nrm1401 [22] Nigg E A. Polo-like kinases: positive regulators of cell division from start to finish[J]. Current Opinion in Cell Biology, 1998, 10(6): 776−783. doi: 10.1016/S0955-0674(98)80121-X [23] 刘镕, 赵琴平, 董惠芬, 等. TGF-β信号传导通路及其生物学功能[J]. 中国病原生物学杂志, 2014, 9(1): 77−83.Liu Rong, Zhao Qinping, Dong Huifen, et al. The TGF-β signaling pathways and their biological functions[J]. Journal of Pathogen Biology, 2014, 9(1): 77−83. [24] Gamer L W, Wolfman N M, Celeste A J, et al. A novel BMP expressed in developing mouse limb, spinal cord, and tail bud is a potent mesoderm inducer in Xenopus embryos[J]. Developmental Biology, 1999, 208(1): 222−232. doi: 10.1006/dbio.1998.9191 [25] Grogg M W, Call M K, Okamoto M, et al. BMP inhibition-driven regulation of six-3 underlies induction of newt lens regeneration[J]. Nature, 2005, 438(7069): 858−862. doi: 10.1038/nature04175 [26] Mashanov V S, Zueva O R, Garcia-Arraras J E, et al. Expression of Wnt9, TCTP, and Bmp1/Tll in sea cucumber visceral regeneration[J]. Gene Expression Patterns, 2012, 12(1/2): 24−35. [27] Han M, Yang Xiangdong, Farrington J E, et al. Digit regeneration is regulated by Msx1 and BMP4 in fetal mice[J]. Development, 2003, 130(21): 5123−5132. doi: 10.1242/dev.00710 [28] Beck C W, Christen B, Barker D, et al. Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles[J]. Mechanisms of Development, 2006, 123(9): 674−688. doi: 10.1016/j.mod.2006.07.001 -





 下载:
下载: